We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 824
From Proteopedia
(Difference between revisions)
| Line 29: | Line 29: | ||
''Note : for a readability purpose the structures enlighten in the Jmol applet will focus only on one SRP54. The same structures are present in the second SRP54M of the dimer.'' | ''Note : for a readability purpose the structures enlighten in the Jmol applet will focus only on one SRP54. The same structures are present in the second SRP54M of the dimer.'' | ||
| - | The secondary structure of Hsrp54M is formed of <scene name='56/568022/Hsrp54m_h1_to_h7/1'>7 alpha helixes (H1 to H7)</scene>. The helices 2 to 7 form the <scene name='56/568022/Hsrp54m_core_and_h1v2/ | + | The secondary structure of Hsrp54M is formed of <scene name='56/568022/Hsrp54m_h1_to_h7/1'>7 alpha helixes (H1 to H7)</scene>. The helices 2 to 7 form the <scene name='56/568022/Hsrp54m_core_and_h1v2/2'>Core structure</scene>, stabilized by hydrophobic, hydrogen and ionic interactions. |
Several residues important to maintain the Core structure were identified. Among them the <scene name='56/568022/Hsrp54m_core_and_h1/2'>Methionine 382,Glutamine 386, Arginine 402 and Arginine 405.</scene> | Several residues important to maintain the Core structure were identified. Among them the <scene name='56/568022/Hsrp54m_core_and_h1/2'>Methionine 382,Glutamine 386, Arginine 402 and Arginine 405.</scene> | ||
| Line 62: | Line 62: | ||
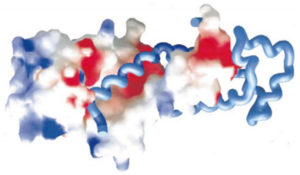
[[Image:Proteopedia charge.png|300px|center|thumb| The helices 4, 5, 6 and 7 surfaces are colored in blue on the spacefill model of SRP54M in this picture. For more details see figure 7 of reference 1.]] | [[Image:Proteopedia charge.png|300px|center|thumb| The helices 4, 5, 6 and 7 surfaces are colored in blue on the spacefill model of SRP54M in this picture. For more details see figure 7 of reference 1.]] | ||
| - | It was also shown that the SRP54M - SRP RNA interaction is highly dependent of the structural integrity of the <scene name='56/568022/Hsrp54m_core_and_h1v2/ | + | It was also shown that the SRP54M - SRP RNA interaction is highly dependent of the structural integrity of the <scene name='56/568022/Hsrp54m_core_and_h1v2/2'>Core structure</scene>. |
Experiments conducted with shorter versions of the M-domain, lacking some aminoacids of the Core, showed a loss in SRP RNA binding activity. | Experiments conducted with shorter versions of the M-domain, lacking some aminoacids of the Core, showed a loss in SRP RNA binding activity. | ||
| Line 68: | Line 68: | ||
This is explained by the fact that theses residues and their lateral chains are inside the core, and can not interact directly with the RNA. They are however important for the conformation of the core. | This is explained by the fact that theses residues and their lateral chains are inside the core, and can not interact directly with the RNA. They are however important for the conformation of the core. | ||
| + | <scene name='56/568022/Hsrp54m_core_and_h1v2/1'>Core structure</scene> | ||
==Related Structures== | ==Related Structures== | ||
Revision as of 13:54, 9 January 2014
Human SRP54 M-domain
| |||||||||||
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |