RuBisCO
From Proteopedia
(→Structural Features) |
|||
| Line 12: | Line 12: | ||
== Large Subunit Structure == | == Large Subunit Structure == | ||
| - | This isolated <scene name='46/463261/Rubisco_lsu_pair/7'>pair of large subunits</scene> shows that each subunit has a large C-terminal lobe and a small N-terminal lobe, and the subunits are arranged head-to-toe (antiparallel). <scene name='46/463261/Rubisco_lsu_pair/5'>Two active sites</scene> are located in the interface of the large subunit pair. The subunits are shown in cartoon with one shown in the secondary structure color scheme. Each active site is occupied by RUBP, which is shown in CPK spacefill. Here is a <scene name='46/463261/Rubisco_lsu_monomer/1'>single large subunit</scene> showing that both lobes contain alpha helices (pink) and beta strands (yellow). The large lobe is dominated by an <scene name='46/463261/Rubisco_lsu_monomer/2'>α-β barrel</scene> (amino acids 166-409), which contributes most of the residues that form | + | This isolated <scene name='46/463261/Rubisco_lsu_pair/7'>pair of large subunits</scene> shows that each subunit has a large C-terminal lobe and a small N-terminal lobe, and the subunits are arranged head-to-toe (antiparallel). <scene name='46/463261/Rubisco_lsu_pair/5'>Two active sites</scene> are located in the interface of the large subunit pair. The subunits are shown in cartoon with one shown in the secondary structure color scheme. Each active site is occupied by RUBP, which is shown in CPK spacefill. Here is a <scene name='46/463261/Rubisco_lsu_monomer/1'>single large subunit</scene> showing that both lobes contain alpha helices (pink) and beta strands (yellow). The large lobe is dominated by an <scene name='46/463261/Rubisco_lsu_monomer/2'>α-β barrel</scene> (amino acids 166-409), which contributes most of the residues that form the active site. One residue from the N-terminal lobe of the adjacent large subunit <scene name='46/463261/Asn123/1'>Asn 123</scene> completes the active site. This scene shows RUBP in spacefill and CPK in one of the active sites in the dimer. Both subunits are shown in transparent cartoon with the α-β barrel is pink and yellow. Asn 123 from the adjacent subunit is in blue spacefill, and residues 121-129 are shown in blue cartoon. This residue does not contribute to catalysis, and it will not be considered further. |
== Active Site Structure == | == Active Site Structure == | ||
| Line 19: | Line 19: | ||
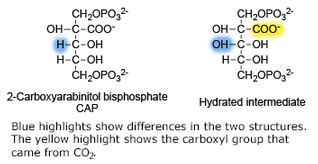
<scene name='46/463261/8ruc_active-site/10'>Residues that are involved in catalysis</scene> are shown shown here in CPK ball & stick. Asp 203 and Glu 204 bind to and position the magnesium ion. The carbamylated lysine residue KCX 201 coordinates Mg<sup>2+</sup> and initiates catalysis by extracting a proton from C3 of RUBP. Note the proximity of the carbamyl group to carbon 3 in this structure. His 294 acts as a catalytic base in the carboxylation step of the mechanism and accepts a proton from the hydroxyl of carbon 3. Mg<sup>2+</sup> is coordinated by six ligands. In addition to oxygen atoms in the three residues already mentioned, the ion binds to two oxygen atoms of RUBP. The 6th ligand is either water or in the carboxylation step it binds the incoming CO<sub>2</sub>. In the structure shown, Mg<sup>2+</sup> is bound to the carboxyl group in CAP that corresponds to the fixed CO<sub>2</sub> in the hydrated intermediate.</StructureSection> | <scene name='46/463261/8ruc_active-site/10'>Residues that are involved in catalysis</scene> are shown shown here in CPK ball & stick. Asp 203 and Glu 204 bind to and position the magnesium ion. The carbamylated lysine residue KCX 201 coordinates Mg<sup>2+</sup> and initiates catalysis by extracting a proton from C3 of RUBP. Note the proximity of the carbamyl group to carbon 3 in this structure. His 294 acts as a catalytic base in the carboxylation step of the mechanism and accepts a proton from the hydroxyl of carbon 3. Mg<sup>2+</sup> is coordinated by six ligands. In addition to oxygen atoms in the three residues already mentioned, the ion binds to two oxygen atoms of RUBP. The 6th ligand is either water or in the carboxylation step it binds the incoming CO<sub>2</sub>. In the structure shown, Mg<sup>2+</sup> is bound to the carboxyl group in CAP that corresponds to the fixed CO<sub>2</sub> in the hydrated intermediate.</StructureSection> | ||
| - | |||
== 3D Structures of RuBisCO == | == 3D Structures of RuBisCO == | ||
Revision as of 14:41, 17 January 2014
Contents |
Ribulose-1,5-bisphosphate carboxylase oxygenase – RuBisCO (RBCO) catalyzes the first step in photosynthetic carbon fixation, and it is the most abundant protein on earth. RBCO can either carboxylate or oxygenate ribulose-1,5-bisphosphate (RUBP) with CO2 or O2, respectively. RBCO from flowering plants consists of eight large subunits and eight small subunits.
Structural Features
| |||||||||||
3D Structures of RuBisCO
Updated February 2013
RuBisCO
3rg6, 1rbl – SeRBCO – Synechococcus elongatus
2ybv - RBCO – Thermosynechococcus elongatus
3qfw - RBCO large subunit – Rhodopseudomonas palustris
1uzh, 1gk8 – CrRBCO – Chlamydomonas reinhardtii
1uw9, 1uwa – CrRBCO (mutant)
1svd – RBCD – Halothiobacillus neapolitanus
1bxn – RBCO – Cupriavidus necator
1aus - spRBCO – spinach
1rba - RrRBCO (mutant) – Rhododpirillum rubrum
5rub - RrRBCO
2wvw – RBCO – Anabena – Cryo EM
2vdh, 2vdi, 2v67, 2v68, 2v63, 2v69, 2v6a - CrRBCO (mutant)
1mlv - pRBCO LSMT – pea
2cxe, 2cwx – PhRBCO - Pyrococcus horikoshii
1uzd – CrRBCO/spRBCO
1geh – TkRBCO – Thermococcus kodakaraensis
1iwa - GpRBCO – Galdieria partita
1tel – RBCO large subunit – Chlorobium tepidum
1rld, 3rub, 3t15, 3zw6, 4rub – tRBCO – tobacco
3thg – RBCO – creosote bush
4hhh – RBCO - pea
RuBisCO complex with inhibitor 2-CABP
3kdn, 3a12 – TkRBCO III + 2-CABP
3kdo, 3a13 - TkRBCO III (mutant) + 2-CABP
1ir2 - CrRBCO + 2-CABP
1upm, 1upp, 1rbo, 3ruc, 8ruc - spRBCO + 2-CABP + cation
1ir1 - spRBCO + 2-CABP + CO2 + Mg
1wdd – rRBCO + 2-CABP – rice
1bwv - GpRBCO + 2-CABP
1rlc - tRBCO + 2-CABP
RuBisCO complex with product
1aa1 – spRBCO + phosphoglycerate
1rus - RrRBCO + phosphoglycerate
RuBisCO complex with substrate
1rcx, 1rxo – spRBCO + ribulose-1,5-bisphosphate
9rub - RrRBCO + ribulose-1,5-bisphosphate
1rsc - SeRBCO + xylulose-1,5-bisphosphate
1rco - spRBCO + xylulose-diol-1,5-bisphosphate
3zxw - SeRBCO + carboxyarabinitol-1,5-bisphosphate
RuBisCO complexes
2h21 – pRBCO LSMT + AdoMet
2h23 - pRBCO LSMT + AdoHcy
2h2e, 1ozv, 1p0y - pRBCO LSMT + AdoMet + lysine
2h2j - pRBCO LSMT + sinefungin
2d69 – PhRBCO + sulfate
2rus - RrRBCO + CO2 + Mg
4f0h – GsRBCO + O2 – Galdieria sulphuraria
4f0k - GsRBCO + CO2 + Mg
4f0m - GsRBCO + Mg
1ej7 – tRBCO + phosphate
3axk – rRBCO + NADP
3axm – rRBCO + 6PG
See Also
Some additional details can be found in Ribulose-1,5-bisphosphate carboxylase/oxygenase.
References
- ↑ Andersson I. Large structures at high resolution: the 1.6 A crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate. J Mol Biol. 1996 May 31;259(1):160-74. PMID:8648644 doi:10.1006/jmbi.1996.0310
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alice Harmon, Joel L. Sussman, Alexander Berchansky