This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Journal:JBIC:24
From Proteopedia
(Difference between revisions)

| Line 9: | Line 9: | ||
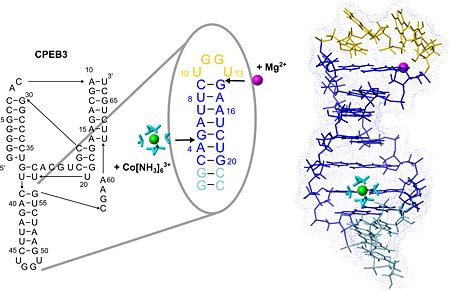
[[Image:JBIC241.jpg|left|450px|thumb|The secondary structure of the CPEB3 ribozyme and the binding sites of Mg2+ and hexammine(III) cobalt that is an NMR-detectable mimic of a hexahydrated Mg2+ ion. P4 hairpin structure is shown with the two suggested metal-ion binding sites.]] | [[Image:JBIC241.jpg|left|450px|thumb|The secondary structure of the CPEB3 ribozyme and the binding sites of Mg2+ and hexammine(III) cobalt that is an NMR-detectable mimic of a hexahydrated Mg2+ ion. P4 hairpin structure is shown with the two suggested metal-ion binding sites.]] | ||
{{Clear}} | {{Clear}} | ||
| - | <scene name='57/573995/Fig/1'> | + | <scene name='57/573995/Fig/1'>P4 contains two well-defined binding sites for Magnesium(II)</scene> (see also image above) ions giving an excellent example of the two different binding modes of Mg(II) to RNA, which are direct coordination and coordination via a shell of water molecules. One of these binding sites probably confers extra stability to the fold of the tetraloop while the other one may play a role in stabilizing the adjacent active site region of the CPEB3 ribozyme. |
| + | The structure of the loop nucleotides is <scene name='57/573995/Fig/2'>exceptionally well defined as indicated by the very good convergence</scene> in this region. <scene name='57/573995/Fig/4'>Stacking interactions</scene> within this new tetraloop structure and a putative hydrogen bond stabilize the hairpin fold which is similar to the AGUU tetraloop structure serving as a protein-binding motif in another RNA. Due to its exposed position and stable fold P4 could serve as an interaction site of the CPEB3 ribozyme for other biomolecules involved in regulating ribozymatic activity. | ||
*<scene name='57/573995/Fig/2'>TextToBeDisplayed</scene> | *<scene name='57/573995/Fig/2'>TextToBeDisplayed</scene> | ||
*<scene name='57/573995/Fig/3'>TextToBeDisplayed</scene> | *<scene name='57/573995/Fig/3'>TextToBeDisplayed</scene> | ||
*<scene name='57/573995/Fig/4'>TextToBeDisplayed</scene> | *<scene name='57/573995/Fig/4'>TextToBeDisplayed</scene> | ||
| - | |||
| - | Stacking interactions within this new tetraloop structure and a putative hydrogen bond stabilize the hairpin fold which is similar to the AGUU tetraloop structure serving as a protein-binding motif in another RNA. | ||
| - | P4 also contains two well-defined binding sites for Magnesium(II) ions giving an excellent example of the two different binding modes of Mg(II) to RNA, which are direct coordination and coordination via a shell of water molecules. One of these binding sites probably confers extra stability to the fold of the tetraloop while the other one may play a role in stabilizing the adjacent active site region of the CPEB3 ribozyme. Due to its exposed position and stable fold P4 could serve as an interaction site of the CPEB3 ribozyme for other biomolecules involved in regulating ribozymatic activity. | ||
</StructureSection> | </StructureSection> | ||
<references/> | <references/> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Revision as of 13:20, 24 March 2014
| |||||||||||
- ↑ REF
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.