We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 911
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
==Hydrolase Information== | ==Hydrolase Information== | ||
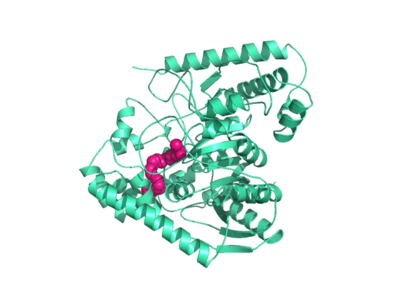
| - | Crystal structures of FAAH show that the enzyme is a homodimer in solution, with each subunit having a mass of 63 kD. The protein's <scene name='57/573125/2vya/8'>twisted Beta sheet core</scene> of 11 strands is surrounded by 24 alpha helices. The enzyme is embedded in the cell membrane to catch the lipid signaling molecules that can diffuse through membranes. The FAAH structure shows an entry channel leading from the lipid bilayer to the enzyme's active site, providing a path for endocannabinoids to enter the hydrolase. In addition, FAAH possesses a channel leading from the active site to the cell's cytoplasm, allowing the release of polar compounds released from lipid cleavage and the entry of water molecules necessary for the FAAH mechanism to proceed. (IMT5) | + | Crystal structures of FAAH show that the enzyme is a homodimer in solution, with each subunit having a mass of 63 kD. The protein's <scene name='57/573125/2vya/8'>twisted Beta sheet core</scene> of 11 strands is surrounded by 24 alpha helices. The enzyme is embedded in the cell <scene name='57/573125/2vya/5'>membrane</scene> to catch the lipid signaling molecules that can diffuse through membranes. The FAAH structure shows an entry channel leading from the lipid bilayer to the enzyme's active site, providing a path for endocannabinoids to enter the hydrolase. In addition, FAAH possesses a channel leading from the active site to the cell's cytoplasm, allowing the release of polar compounds released from lipid cleavage and the entry of water molecules necessary for the FAAH mechanism to proceed. (IMT5) |
This hydrolase has a membrane binding cap, a <scene name='57/573125/2vya/7'>helix-turn-helix motif</scene> consisting of alpha helices 18 and 19. These helices present hydrophobic amino acid residues that likely help FAAH interact with the hydrophobic region of the lipid bilayer. (IMT5) | This hydrolase has a membrane binding cap, a <scene name='57/573125/2vya/7'>helix-turn-helix motif</scene> consisting of alpha helices 18 and 19. These helices present hydrophobic amino acid residues that likely help FAAH interact with the hydrophobic region of the lipid bilayer. (IMT5) | ||
Revision as of 13:02, 25 March 2014
| This Sandbox is Reserved from Jan 06, 2014, through Aug 22, 2014 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 911 through Sandbox Reserved 922. |
To get started:
More help: Help:Editing |
| |||||||||||
Applications
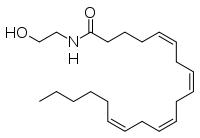
The human nervous system has several types of chemical messengers, including amino acids, lipids, peptide hormones, and monoamines. (IMT5) FAAH primarily degrades anandamide (AEA), a naturally-occurring signaling lipid that functions in the brain. AEA brings pain relief to the body. Inhibiting FAAH would likely sustain AEA signaling, leading to prolonged pain relief and decreased inflammation. (2YVA)