This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox reserved 916
From Proteopedia
(Difference between revisions)
| Line 21: | Line 21: | ||
====Binding==== | ====Binding==== | ||
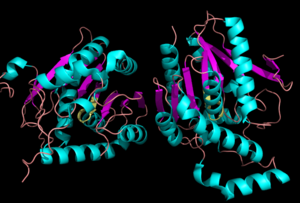
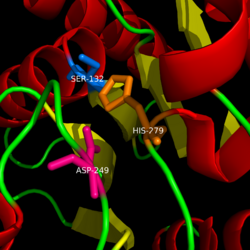
| - | The <scene name='58/580299/Ligand_binding_pocket/2'> ligand binding pocket </scene> of MGL has a large hydrophobic region with a polar bottom (Bertrand et al. 2010). Bertrand found that in MGL the binding pocket is not adjusted to the ligands shape. 2-archidonylglycerol are ligands for cannabinoid receptors (Clemente et al. 2012). Inhibition of MGL leads to increase in 2-AG levels since AG is broken down by MGL (Clemente et al. 2012). Through covalent interactions with a Cys residue, NAM, one of the many possible inhibitors, is able to inhibit MGL (Bertrand et al. 2010). | ||
| Line 27: | Line 26: | ||
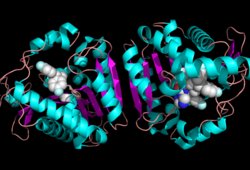
[[Image:Overall_ligand.png|left|250px|thumb|Ligand within the Overall Structure of MGL]] | [[Image:Overall_ligand.png|left|250px|thumb|Ligand within the Overall Structure of MGL]] | ||
<scene name='58/580298/Ligand/1'>Ligand binding site</scene> | <scene name='58/580298/Ligand/1'>Ligand binding site</scene> | ||
| - | + | The <scene name='58/580299/Ligand_binding_pocket_take_3/1'> ligand binding pocket </scene> of MGL has a large hydrophobic region with a polar bottom (Bertrand et al. 2010). Bertrand found that in MGL the binding pocket is not adjusted to the ligands shape. 2-archidonylglycerol are ligands for cannabinoid receptors (Clemente et al. 2012). Inhibition of MGL leads to increase in 2-AG levels since AG is broken down by MGL (Clemente et al. 2012). Through covalent interactions with a Cys residue, NAM, one of the many possible inhibitors, is able to inhibit MGL (Bertrand et al. 2010). | |
==Overall Reaction== | ==Overall Reaction== | ||
Revision as of 13:35, 25 March 2014
Monoglyceride Lipase (MGL)
| |||||||||||