We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox reserved 915
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
In Peripheral tissues, EC is active in autonomic nervous system. EC affects processes such as learning, motor control, cognition, and pain (Taschler et al. 2011). EC is also able to regulate lipid metabolism and food intake (Taschler et al. 2011). | In Peripheral tissues, EC is active in autonomic nervous system. EC affects processes such as learning, motor control, cognition, and pain (Taschler et al. 2011). EC is also able to regulate lipid metabolism and food intake (Taschler et al. 2011). | ||
Taschler et al. looked at the role of MGL in energy metabolism, finding that MGL deficiency in animals led to the buildup of 2-AG (Taschler et al. 2011). | Taschler et al. looked at the role of MGL in energy metabolism, finding that MGL deficiency in animals led to the buildup of 2-AG (Taschler et al. 2011). | ||
| + | ===Inhibition of MGL=== | ||
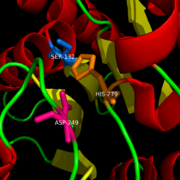
| + | Studies have shown that NAM inhibits MGL by reacting with the amino acid Cys252. This Cysteine is buried in the active site near the catalytic serine. This inhibition can be explained by a steric clash between the inhibitor and the natural ligand. There is also a possibility of conformational changes upon the binding of the cysteine that would lead to an inactive form of MGL. | ||
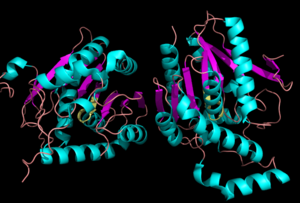
==Structure== | ==Structure== | ||
Representation of the <scene name='58/580298/Overall_structure/3'>Overall Structure</scene> | Representation of the <scene name='58/580298/Overall_structure/3'>Overall Structure</scene> | ||
| Line 17: | Line 19: | ||
====Binding==== | ====Binding==== | ||
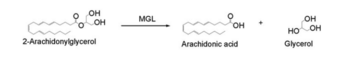
2-AG binds to the catalytic triad and is hydrolyzed. The structure of 2-AG contains a long and flexible aliphatic chain and a polar head that is cleaved. 2-AG is broken down into arachidonic acid and glycerol which makes 2-AG inactive. '''See Overall Reaction.''' | 2-AG binds to the catalytic triad and is hydrolyzed. The structure of 2-AG contains a long and flexible aliphatic chain and a polar head that is cleaved. 2-AG is broken down into arachidonic acid and glycerol which makes 2-AG inactive. '''See Overall Reaction.''' | ||
| - | ===Inhibition=== | + | ===Inhibition of Catalytic Triad=== |
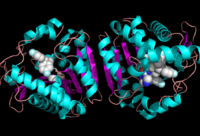
Research on MGL is being geared towards inhibiting 2-AG from binding to the catalytic triad and being hydrolyzed. The binding of 2-AG to the catalytic triad can not be inhibited, but it can be extracted before being hydrolyzed. MPD (2-methyl-pentane-2,4-diol)is located at the end of the tunnel where the catalytic triad is at and the tunnel is filled with MPD molecules. MPD being in the same vicinity will extract 2-AG from the triad and the MPD molecule will sit in there in place of 2-AG. This natural inhibition phenomenon is known as interfacial activation. | Research on MGL is being geared towards inhibiting 2-AG from binding to the catalytic triad and being hydrolyzed. The binding of 2-AG to the catalytic triad can not be inhibited, but it can be extracted before being hydrolyzed. MPD (2-methyl-pentane-2,4-diol)is located at the end of the tunnel where the catalytic triad is at and the tunnel is filled with MPD molecules. MPD being in the same vicinity will extract 2-AG from the triad and the MPD molecule will sit in there in place of 2-AG. This natural inhibition phenomenon is known as interfacial activation. | ||
| Line 36: | Line 38: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | 1) Bertrand, T., F. Augé, J. Houtmann, A. Rak, F. Vallée, V. Mikol, P.f. Berne, N. Michot, D. Cheuret, C. Hoornaert, and M. Mathieu. "Structural Basis for Human Monoglyceride Lipase Inhibition." Journal of Molecular Biology 396.3 (2010): 663-73. | ||
| + | |||
== External links == | == External links == | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 19:54, 30 March 2014
Monoglyceride Lipase (MGL)
| |||||||||||