Sandbox Reserved 918
From Proteopedia
| Line 16: | Line 16: | ||
<StructureSection load='1X70' size='350' frame='true' align='right' caption='Biological Dimer of DPP IV' scene='57/573132/1x70_basic_dimer/1'> | <StructureSection load='1X70' size='350' frame='true' align='right' caption='Biological Dimer of DPP IV' scene='57/573132/1x70_basic_dimer/1'> | ||
===Binding Pocket=== | ===Binding Pocket=== | ||
| - | The specificity of the DPP IV in its ability to discern the proline from other amino acids can be seen in the binding pocket where two glutamates, <scene name='57/573132/1x70_glutamates/3'>Glu205-Glu206</scene> orient the substrate allowing only small residues like proline or alanine to fit. The [http://en.wikipedia.org/wiki/Glutamic_acid glutamates] form a [http://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) salt bridge] with the N-terminus, positioning the substrate so that only two amino acids can fit into position for | + | The specificity of the DPP IV in its ability to discern the proline from other amino acids can be seen in the binding pocket where two glutamates, <scene name='57/573132/1x70_glutamates/3'>Glu205-Glu206</scene> orient the substrate allowing only small residues like proline or alanine to fit. The [http://en.wikipedia.org/wiki/Glutamic_acid glutamates] form a [http://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) salt bridge] with the N-terminus, positioning the substrate so that only two amino acids can fit into position for hydrolysis. <ref name="Gorrell">PMID: 15584901</ref> Examples of DPP IV substrates with alanine or proline at their N-terminus are: |
{| class="wikitable" style="text-align:center; width:550px; height:200px;" | {| class="wikitable" style="text-align:center; width:550px; height:200px;" | ||
|+ | |+ | ||
| Line 42: | Line 42: | ||
===Active Site=== | ===Active Site=== | ||
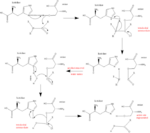
| - | These substrates, along with many others, are cleaved by DPP IV at its active site containing a [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] composed of <scene name='57/573132/1x70_catalytictriad/1'>Ser630, His740, and Asp708</scene>. The substrate shown is a DPP IV | + | These substrates, along with many others, are cleaved by DPP IV at its active site containing a [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] composed of <scene name='57/573132/1x70_catalytictriad/1'>Ser630, His740, and Asp708</scene>. The substrate shown is a DPP IV inhibitor. This Serine-Histidine-Asparatate motif, best known in the enzyme [http://en.wikipedia.org/wiki/Chymotrypsin chymotrypsin], uses acid-base chemistry to facilitate the binding, cleaving, and release of the given substrate. The mechanism of the reaction is as follows: |
<div style="text-align: left;"> | <div style="text-align: left;"> | ||
# Substrate binds to enzyme and carbonyl carbon is positioned by [http://en.wikipedia.org/wiki/Active_site active site]. | # Substrate binds to enzyme and carbonyl carbon is positioned by [http://en.wikipedia.org/wiki/Active_site active site]. | ||
| - | # | + | # Histidine, via hydrogen bond with asparatate, becomes more [http://en.wikipedia.org/wiki/Electronegativity electronegative] and therefore readily accepts the hydrogen from the -OH group on serine, making it [http://en.wikipedia.org/wiki/Nucleophile nucleophilic]. [[Image:Serine_protease_mechanism_by_snellios.png|right|thumb|150px|<font size=".8"><div style="text-align: center;"> Arrow Pushing Mechanism </div></font>]] |
# The nucleophilic serine attacks the [http://en.wikipedia.org/wiki/Carbonyl carbonyl] carbon, generating a [http://goldbook.iupac.org/T06289.html tetrahedral intermediate] (as seen in the [http://en.wikipedia.org/wiki/Arrow_pushing arrow pushing mechanism]). | # The nucleophilic serine attacks the [http://en.wikipedia.org/wiki/Carbonyl carbonyl] carbon, generating a [http://goldbook.iupac.org/T06289.html tetrahedral intermediate] (as seen in the [http://en.wikipedia.org/wiki/Arrow_pushing arrow pushing mechanism]). | ||
| - | # The peptide bond is cleaved and the electrons from it move to attack the hydrogen on the | + | # The peptide bond is cleaved and the electrons from it move to attack the hydrogen on the histidine. This half of the substrate now dissociates, leaving the other half still bound to serine. |
| - | # | + | # Histidine then deprotonates a water molecule, forming a nucleophilic [http://en.wikipedia.org/wiki/Hydroxide hydroxide group] that attacks the serine-bound carbonyl carbon. |
# As the electrons move from the oxygen back down to reform the double bond, serine [http://en.wikipedia.org/wiki/Dissociation_(chemistry) dissociates] as a leaving group and the other half of the substrate dissociates from the enzyme. | # As the electrons move from the oxygen back down to reform the double bond, serine [http://en.wikipedia.org/wiki/Dissociation_(chemistry) dissociates] as a leaving group and the other half of the substrate dissociates from the enzyme. | ||
| - | # The negative oxygen on serine readily accepts the hydrogen from | + | # The negative oxygen on serine readily accepts the hydrogen from histidine and in doing so regenerates the active site of the enzyme. |
</div> | </div> | ||
Revision as of 17:51, 31 March 2014
| This Sandbox is Reserved from Jan 06, 2014, through Aug 22, 2014 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 911 through Sandbox Reserved 922. |
To get started:
More help: Help:Editing |
Contents |
Dipeptidyl Peptidase IV
Introduction
Dipeptidyl Peptidase IV (commonly abbreviated as DPP IV) is a regulatory protease and binding glycoprotein that carries out numerous functions in humans making it a prime candidate for medicinal and pharmaceutical research. DPP IV, discovered by V.K. Hopsu-Havu and G.G. Glenner in homogenized rat liver tissue[1], was originally believed to serve a specific role in breaking 2-Naphthylamine off of Gly-Pro-2-napthylamide, hence its original name glycylproline napthylamidase. However, further research into the specificity of DPP IV eventually showed that it serves a more generic function as a hydrolase (a serine exopeptidase), breaking N-terminal Xaa-Pro bonds (though it can alΌso catalyze alanine bonds). DPP IV is the founding member of the DPP-IV and/or structure homologue (DASH) family, who all share this serine protease catalysis of post-proline peptide bonds. [2] These penultimate prolines of the N-terminus are known for their ability to resist attacks from most proteases and also induce a conformational change of their respective proteins. Also, DPP IV serves as a binding glycoprotein on the membrane of cells, binding ligands such as adenosine deaminase with high affinity.[3] Though this interaction has no known significance as of yet, DPP IV and its ability to catalyze N-terminal prolines gives it a unique specificity and target for pharmaceutical companies to take advantage of. [1]
Structure
| |||||||||||
Medical Relevancy
References
- ↑ 1.0 1.1 Mentlein R. Dipeptidyl-peptidase IV (CD26)--role in the inactivation of regulatory peptides. Regul Pept. 1999 Nov 30;85(1):9-24. PMID:10588446
- ↑ Lankas GR, Leiting B, Roy RS, Eiermann GJ, Beconi MG, Biftu T, Chan CC, Edmondson S, Feeney WP, He H, Ippolito DE, Kim D, Lyons KA, Ok HO, Patel RA, Petrov AN, Pryor KA, Qian X, Reigle L, Woods A, Wu JK, Zaller D, Zhang X, Zhu L, Weber AE, Thornberry NA. Dipeptidyl peptidase IV inhibition for the treatment of type 2 diabetes: potential importance of selectivity over dipeptidyl peptidases 8 and 9. Diabetes. 2005 Oct;54(10):2988-94. PMID:16186403
- ↑ 3.0 3.1 3.2 3.3 3.4 Gorrell MD. Dipeptidyl peptidase IV and related enzymes in cell biology and liver disorders. Clin Sci (Lond). 2005 Apr;108(4):277-92. PMID:15584901 doi:http://dx.doi.org/10.1042/CS20040302