We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Daud Akhtar/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
===Substrate(Glucose-6-Phosphate) Binding Domain=== | ===Substrate(Glucose-6-Phosphate) Binding Domain=== | ||
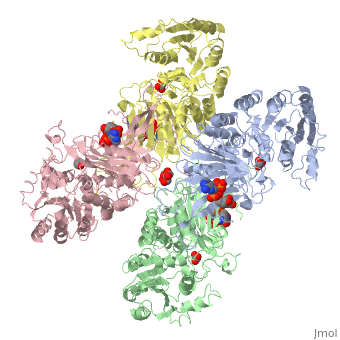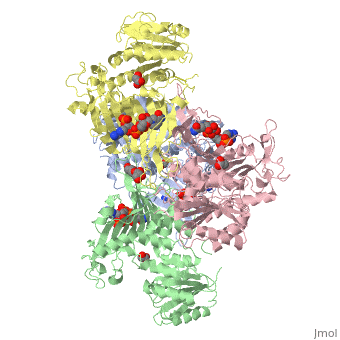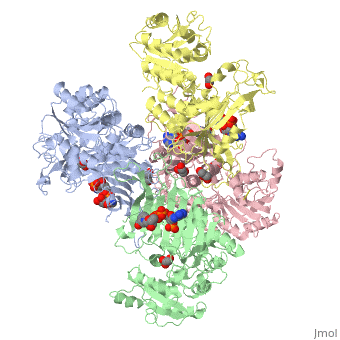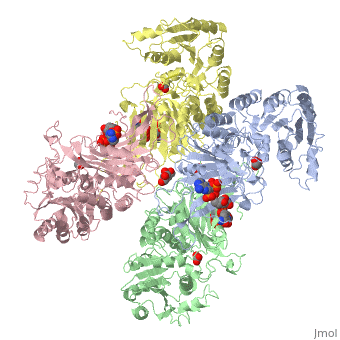
| - | The second domain is the<scene name='58/580852/G6p_binding_site/1'> substrate(G6P) binding domain(shown in the A monomer)</scene> consists of both α-helices and β-sheets with nine distinct anti-parallel β-sheets. The active site in this domain corresponds to the site for substrate (G6P) and is characterized by the highly conserved amino acid residues RIDHYLGK (amino acids 198-205). In regards to the overall dimer stability, Au (2000) illustrated that the second domain plays a role in dimerization. Specifically, within each subunit a NADP+ is buried in a structural moiety between the β-sheet and the C-terminus of the first domain. In this structural moiety, NADP+ does not act as a coenzyme but rather as a stabilizer. Specifically, the adenine and nicotinamide form hydrophobic interactions with Tyr503 and Arg487, and Trp509 and Tyr 401 respectively. The amide portion of the NADP+ interacts with Asp421 and Arg393 and the bisphosphate interacts with Arg370 through hydrogen bonding. The two monomers are linked together with the amino acids Lys275 and Lys290 of one monomer forming salt bridges with Glu347 and Glu287 <ref name="1qki" />. | + | The second domain is the<scene name='58/580852/G6p_binding_site/1'> substrate(G6P) binding domain(shown in the A monomer)</scene> consists of both α-helices and β-sheets with nine distinct anti-parallel β-sheets. The active site in this domain corresponds to the site for substrate (G6P) and is characterized by the highly conserved amino acid residues RIDHYLGK (amino acids 198-205). In regards to the overall dimer stability, Au (2000) illustrated that the second domain plays a role in dimerization. Specifically, within each subunit a NADP+ is buried in a structural moiety between the β-sheet and the C-terminus of the first domain. In this structural moiety, NADP+ does not act as a coenzyme but rather as a stabilizer. Specifically, the adenine and nicotinamide form hydrophobic interactions with Tyr503 and Arg487, and Trp509 and Tyr 401 respectively. The amide portion of the NADP+ interacts with Asp421 and Arg393 and the bisphosphate interacts with Arg370 through hydrogen bonding. The two monomers are linked together with the amino acids<scene name='58/580852/Monomer_links/1'> Lys275 and Lys290 of one monomer forming salt bridges with Glu347 and Glu287</scene> <ref name="1qki" />. |
===Substrate Binding and Catalytic Mechanism=== | ===Substrate Binding and Catalytic Mechanism=== | ||
Revision as of 01:26, 1 April 2014
Glucose-6-Phosphate Dehydrogenase(G6PD)
| |||||||||||
References
- ↑ Salati LM, Amir-Ahmady B. Dietary regulation of expression of glucose-6-phosphate dehydrogenase. Annu Rev Nutr. 2001;21:121-40. PMID:11375432 doi:http://dx.doi.org/10.1146/annurev.nutr.21.1.121
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Au SW, Gover S, Lam VM, Adams MJ. Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency. Structure. 2000 Mar 15;8(3):293-303. PMID:10745013
- ↑ Kotaka M, Gover S, Vandeputte-Rutten L, Au SW, Lam VM, Adams MJ. Structural studies of glucose-6-phosphate and NADP+ binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2005 May;61(Pt 5):495-504. Epub 2005, Apr 20. PMID:15858258 doi:http://dx.doi.org/10.1107/S0907444905002350
- ↑ Corpas FJ, Barroso JB, Sandalio LM, Distefano S, Palma JM, Lupianez JA, Del Rio LA. A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem J. 1998 Mar 1;330 ( Pt 2):777-84. PMID:9480890
- ↑ Au SW, Naylor CE, Gover S, Vandeputte-Rutten L, Scopes DA, Mason PJ, Luzzatto L, Lam VM, Adams MJ. Solution of the structure of tetrameric human glucose 6-phosphate dehydrogenase by molecular replacement. Acta Crystallogr D Biol Crystallogr. 1999 Apr;55(Pt 4):826-34. PMID:10089300
- ↑ Bhadbhade MM, Adams MJ, Flynn TG, Levy HR. Sequence identity between a lysine-containing peptide from Leuconostoc mesenteroides glucose-6-phosphate dehydrogenase and an active site peptide from human erythrocyte glucose-6-phosphate dehydrogenase. FEBS Lett. 1987 Jan 26;211(2):243-6. PMID:3100332
- ↑ Cosgrove MS, Naylor C, Paludan S, Adams MJ, Levy HR. On the mechanism of the reaction catalyzed by glucose 6-phosphate dehydrogenase. Biochemistry. 1998 Mar 3;37(9):2759-67. PMID:9485426 doi:10.1021/bi972069y
- ↑ Ramos KL, Colquhoun A. Protective role of glucose-6-phosphate dehydrogenase activity in the metabolic response of C6 rat glioma cells to polyunsaturated fatty acid exposure. Glia. 2003 Aug;43(2):149-66. PMID:12838507 doi:http://dx.doi.org/10.1002/glia.10246
- ↑ Tian WN, Braunstein LD, Pang J, Stuhlmeier KM, Xi QC, Tian X, Stanton RC. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J Biol Chem. 1998 Apr 24;273(17):10609-17. PMID:9553122
- ↑ Scott MD, Zuo L, Lubin BH, Chiu DT. NADPH, not glutathione, status modulates oxidant sensitivity in normal and glucose-6-phosphate dehydrogenase-deficient erythrocytes. Blood. 1991 May 1;77(9):2059-64. PMID:2018843
- ↑ Scott MD, Zuo L, Lubin BH, Chiu DT. NADPH, not glutathione, status modulates oxidant sensitivity in normal and glucose-6-phosphate dehydrogenase-deficient erythrocytes. Blood. 1991 May 1;77(9):2059-64. PMID:2018843
- ↑ . Glucose-6-phosphate dehydrogenase deficiency. WHO Working Group. Bull World Health Organ. 1989;67(6):601-11. PMID:2633878
- ↑ Manganelli G, Masullo U, Passarelli S, Filosa S. Glucose-6-phosphate dehydrogenase deficiency: disadvantages and possible benefits. Cardiovasc Hematol Disord Drug Targets. 2013 Mar 1;13(1):73-82. PMID:23534950
- ↑ Beutler E. Glucose-6-phosphate dehydrogenase deficiency. N Engl J Med. 1991 Jan 17;324(3):169-74. PMID:1984194 doi:http://dx.doi.org/10.1056/NEJM199101173240306