We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Daud Akhtar/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
===Substrate(Glucose-6-Phosphate) Binding Domain=== | ===Substrate(Glucose-6-Phosphate) Binding Domain=== | ||
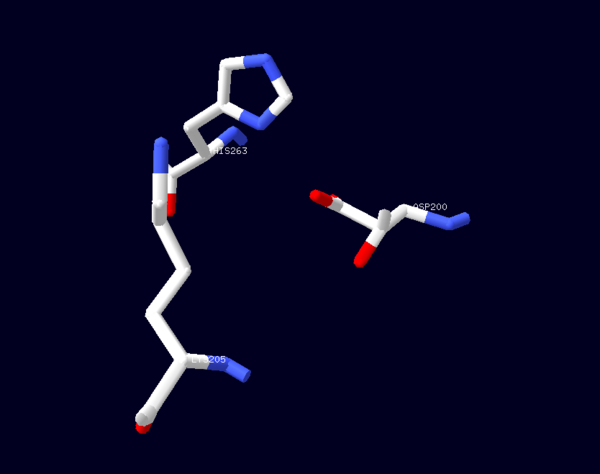
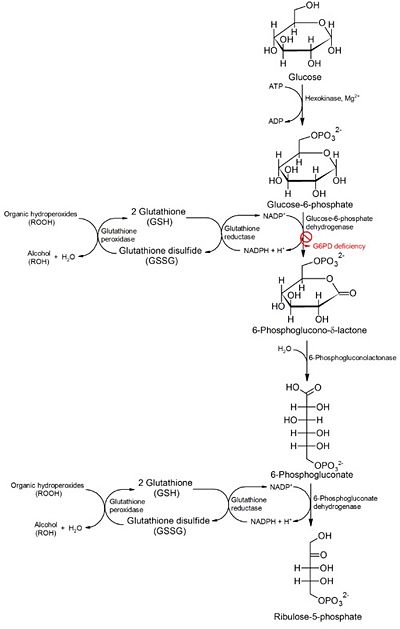
| - | The second domain is the<scene name='58/580852/G6p_binding_site/1'> substrate(G6P) binding domain(shown in the A monomer)</scene> consists of both α-helices and β-sheets with nine distinct anti-parallel β-sheets. The active site in this domain corresponds to the site for substrate (G6P) and is characterized by the highly conserved amino acid residues RIDHYLGK (amino acids 198-205). In regards to the overall dimer stability, Au (2000) illustrated that the second domain plays a role in dimerization. Specifically, within each subunit a NADP+ is buried in a structural moiety between the β-sheet and the C-terminus of the first domain. In this structural moiety, NADP+ does not act as a coenzyme but rather as a stabilizer. Specifically, the adenine and nicotinamide form hydrophobic interactions with Tyr503 and Arg487, and Trp509 and Tyr 401 respectively. The amide portion of the NADP+ interacts with Asp421 and Arg393 and the bisphosphate interacts with Arg370 through hydrogen bonding. The two monomers are linked together with the amino acids<scene name='58/580852/Monomer_links/ | + | The second domain is the<scene name='58/580852/G6p_binding_site/1'> substrate(G6P) binding domain(shown in the A monomer)</scene> consists of both α-helices and β-sheets with nine distinct anti-parallel β-sheets. The active site in this domain corresponds to the site for substrate (G6P) and is characterized by the highly conserved amino acid residues RIDHYLGK (amino acids 198-205). In regards to the overall dimer stability, Au (2000) illustrated that the second domain plays a role in dimerization. Specifically, within each subunit a NADP+ is buried in a structural moiety between the β-sheet and the C-terminus of the first domain. In this structural moiety, NADP+ does not act as a coenzyme but rather as a stabilizer. Specifically, the adenine and nicotinamide form hydrophobic interactions with Tyr503 and Arg487, and Trp509 and Tyr 401 respectively. The amide portion of the NADP+ interacts with Asp421 and Arg393 and the bisphosphate interacts with Arg370 through hydrogen bonding. The two monomers are linked together with the amino acids<scene name='58/580852/Monomer_links/2'> Lys275 and Lys290 of one monomer forming salt bridges with Glu347 and Glu287</scene><ref name="1qki" />. |
===Substrate Binding and Catalytic Mechanism=== | ===Substrate Binding and Catalytic Mechanism=== | ||
Revision as of 01:44, 1 April 2014
Glucose-6-Phosphate Dehydrogenase(G6PD)
| |||||||||||
References
- ↑ Salati LM, Amir-Ahmady B. Dietary regulation of expression of glucose-6-phosphate dehydrogenase. Annu Rev Nutr. 2001;21:121-40. PMID:11375432 doi:http://dx.doi.org/10.1146/annurev.nutr.21.1.121
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Au SW, Gover S, Lam VM, Adams MJ. Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency. Structure. 2000 Mar 15;8(3):293-303. PMID:10745013
- ↑ Kotaka M, Gover S, Vandeputte-Rutten L, Au SW, Lam VM, Adams MJ. Structural studies of glucose-6-phosphate and NADP+ binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2005 May;61(Pt 5):495-504. Epub 2005, Apr 20. PMID:15858258 doi:http://dx.doi.org/10.1107/S0907444905002350
- ↑ Corpas FJ, Barroso JB, Sandalio LM, Distefano S, Palma JM, Lupianez JA, Del Rio LA. A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem J. 1998 Mar 1;330 ( Pt 2):777-84. PMID:9480890
- ↑ Au SW, Naylor CE, Gover S, Vandeputte-Rutten L, Scopes DA, Mason PJ, Luzzatto L, Lam VM, Adams MJ. Solution of the structure of tetrameric human glucose 6-phosphate dehydrogenase by molecular replacement. Acta Crystallogr D Biol Crystallogr. 1999 Apr;55(Pt 4):826-34. PMID:10089300
- ↑ Bhadbhade MM, Adams MJ, Flynn TG, Levy HR. Sequence identity between a lysine-containing peptide from Leuconostoc mesenteroides glucose-6-phosphate dehydrogenase and an active site peptide from human erythrocyte glucose-6-phosphate dehydrogenase. FEBS Lett. 1987 Jan 26;211(2):243-6. PMID:3100332
- ↑ Cosgrove MS, Naylor C, Paludan S, Adams MJ, Levy HR. On the mechanism of the reaction catalyzed by glucose 6-phosphate dehydrogenase. Biochemistry. 1998 Mar 3;37(9):2759-67. PMID:9485426 doi:10.1021/bi972069y
- ↑ Ramos KL, Colquhoun A. Protective role of glucose-6-phosphate dehydrogenase activity in the metabolic response of C6 rat glioma cells to polyunsaturated fatty acid exposure. Glia. 2003 Aug;43(2):149-66. PMID:12838507 doi:http://dx.doi.org/10.1002/glia.10246
- ↑ Tian WN, Braunstein LD, Pang J, Stuhlmeier KM, Xi QC, Tian X, Stanton RC. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J Biol Chem. 1998 Apr 24;273(17):10609-17. PMID:9553122
- ↑ Scott MD, Zuo L, Lubin BH, Chiu DT. NADPH, not glutathione, status modulates oxidant sensitivity in normal and glucose-6-phosphate dehydrogenase-deficient erythrocytes. Blood. 1991 May 1;77(9):2059-64. PMID:2018843
- ↑ Scott MD, Zuo L, Lubin BH, Chiu DT. NADPH, not glutathione, status modulates oxidant sensitivity in normal and glucose-6-phosphate dehydrogenase-deficient erythrocytes. Blood. 1991 May 1;77(9):2059-64. PMID:2018843
- ↑ . Glucose-6-phosphate dehydrogenase deficiency. WHO Working Group. Bull World Health Organ. 1989;67(6):601-11. PMID:2633878
- ↑ Manganelli G, Masullo U, Passarelli S, Filosa S. Glucose-6-phosphate dehydrogenase deficiency: disadvantages and possible benefits. Cardiovasc Hematol Disord Drug Targets. 2013 Mar 1;13(1):73-82. PMID:23534950
- ↑ Beutler E. Glucose-6-phosphate dehydrogenase deficiency. N Engl J Med. 1991 Jan 17;324(3):169-74. PMID:1984194 doi:http://dx.doi.org/10.1056/NEJM199101173240306