Background
Monoglyceride lipase is part of the α/β hydrolase family, having a Ser-His-Asp catalytic triad (Celemnte et al. 2012). This enzyme is present in most cells, providing the rate limiting step for MG (Taschler et al 2011). MGL terminates the signaling of a primary endocannabinoid, 2-AG (Savinainen et al 2010). MGL is the main enzyme respondsible for hydrolyzing 2-arachidonoylglycerol into arachidonic acid and glycerol in vivo [1]. One of the key features of MGL is the hydrophobic tunnel, which has been suggested to provide a model for drug research.
Metabolic Role
Monoglyceride lipase is able to hydrolyze monoacylglycerols into fatty acids and glycerol, which are able to then be used for energy production (Taschler et al. 2011). MGL degrades sn-1 and 2-MG at identical specific rates as a part of its metabolic role (Taschler et al. 2011).
Component of Endocannabinoid System
MGL degrades 2-arachidonoyl glycerol (2-AG). 2-AG is commonly classified as an endocannabinoid. In the brain endocannabinoids are released from postsynaptic neurons, causing the retrograde suppression of synaptic transmission (Taschler et al. 2011).
In Peripheral tissues, EC is active in autonomic nervous system. EC affects processes such as learning, motor control, cognition, and pain (Taschler et al. 2011). EC is also able to regulate lipid metabolism and food intake (Taschler et al. 2011).
Taschler et al. looked at the role of MGL in energy metabolism, finding that MGL deficiency in animals led to the buildup of 2-AG (Taschler et al. 2011).
Inhibition of MGL
Studies have shown that N-arachidonylmaleimide (NAM) inhibits MGL by reacting with the amino acid . This Cysteine is buried in the active site near the catalytic serine. This inhibition can be explained by a steric clash between the inhibitor and the natural ligand. There is also a possibility of conformational changes upon the binding of the cysteine that would lead to an inactive form of MGL.
MGL is also inhibited by being in complex with SAR629 that is covalently bound to the catalytic Ser132. SAR629 adopts a Y shape and interacts with the MGL by hydrophobic interactions, with a few polar interactions as well. With SAR629 interacting with the catalytic triad it inhibits the triad from breaking down 2-AG and it interacting with MGL makes it inactive [2].
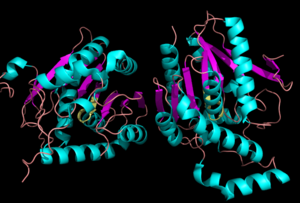
Structure
Representation of the of MGL.
MGL has eight-stranded β-sheet protein fold with seven parallel and one (Bertrand et al. 2010). The β-sheets are surrounded by α-helices. The combination of the α-helices and β-sheets are able to provide a stable scaffold for the active sites within MGL (Bertrand et al. 2010). Within the main domain of MGL is the conserved catalytic triad (Bertrand et al. 2010).
Catalytic triad
MGL has a classic that contains Ser-His-Asp (figure 1). The triad was found using site-directed mutagenesis of each individual residue and this experiment confirmed that these amino acid residues are catalytically essential. The catalytic triad is located in the binding pocket buried at the bottom of it in the oxyanion hole connected by a water molecule. This triad has a natural attraction to Endocannabinoids, specifically 2-arachidonylglycerol (2-AG). 2-AG contributes to brain signals (neurons) to suppress the pain pathways when a patient is feeling depressed or suffering from any type of pain.
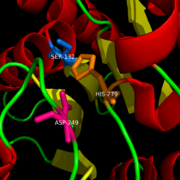
Figure 1: Catalytic Triad of MGL structure
Binding
2-AG binds to the catalytic triad and is hydrolyzed. The structure of 2-AG contains a long and flexible aliphatic chain and a polar head that is cleaved. 2-AG is broken down into arachidonic acid and glycerol which makes 2-AG inactive. See Overall Reaction.
Inhibition of Catalytic Triad
Research on MGL is being geared towards inhibiting 2-AG from binding to the catalytic triad and being hydrolyzed. The binding of 2-AG to the catalytic triad can not be inhibited, but it can be extracted before being hydrolyzed. MPD (2-methyl-pentane-2,4-diol)is located at the end of the tunnel where the catalytic triad is at and the tunnel is filled with MPD molecules. MPD being in the same vicinity will extract 2-AG from the triad and the MPD molecule will sit in there in place of 2-AG. This natural inhibition phenomenon is known as interfacial activation. Inhibition of MGL leads to increase in 2-AG levels since AG is broken down by MGL (Clemente et al. 2012). Through covalent interactions with a Cys residue, NAM, one of the many possible inhibitors, is able to inhibit MGL (Bertrand et al. 2010).
Ligand Binding Site
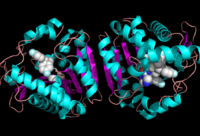
Ligand within the Overall Structure of MGL
The of MGL has a large hydrophobic region with a polar bottom (Bertrand et al. 2010). The entrance of the binding pocket for MGL contains a lid, which is very flexible (Bertrand et al. 2010). The binding pocket or tunnel within MGL matches with the overall structure of 2-AG, with 2-AG's polar head being cleaved by the catalytic triad (Bertrand et al. 2010). Bertrand found that in MGL the binding pocket is not adjusted to the ligand's shape. However, the main movements of MGL associated with ligand binding involved the lid region (Bertrand et al. 2010). When 2-AG and its isomer 1(3)-AG bind to MGL, the hydrophobic chain is first aligned with the left part of the binding pocket. The carbonyl is then hydrogen bonded to Ala61 (Bertrand et al. 2010). The polar head group of the ligand is then fixed by three hydrogen bonds (Bertrand et al. 2010). As a result, future research is looking into the large lipophilic portion of the binding pocket for designing selective inhibitors.
Overall Reaction
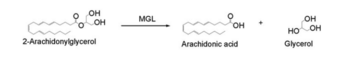
Overall reaction representing the hydrolysis of 2-AG by MGL
In this reaction 2-AG binds to the catalytic triad in the oxyanion hole in the active site. In the oxyanion holes the oxygen of the substrate is stabilized by two nitrogen atoms during the transition step of the catalytic reaction. The triad activates the nucleophilic serine and cleaves the ester bond of 2-AG that is being stabilized by its carbonyl group that is attached to the oxyanion hole. The glycerol molecule is released and it might diffuse to the narrow "exit hole", while the arachidonic acid would diffuse back to the top of the tunnel and leave the protein.
Additional Resources
Serine hydrolases
Lipases
Binding Pockets
References
- ↑ [ Bertrand, T., F. Augé, J. Houtmann, A. Rak, F. Vallée, V. Mikol, P.f. Berne, N. Michot, D. Cheuret, C. Hoornaert, and M. Mathieu. "Structural Basis for Human Monoglyceride Lipase Inhibition." Journal of Molecular Biology 396.3 (2010): 663-73.]
- ↑ [1]
1) Bertrand, T., F. Augé, J. Houtmann, A. Rak, F. Vallée, V. Mikol, P.f. Berne, N. Michot, D. Cheuret, C. Hoornaert, and M. Mathieu. "Structural Basis for Human Monoglyceride Lipase Inhibition." Journal of Molecular Biology 396.3 (2010): 663-73.
2) Clemente, J. C., E. Nulton, M. Nelen, M. J. Todd, D. Maguire, C. Schalk-Hihi, L. C. Kuo, S.-P. Zhang, C. M. Flores, and J. K. Kranz. "Screening and Characterization of Human Monoglyceride Lipase Active Site Inhibitors Using Orthogonal Binding and Functional Assays." Journal of Biomolecular Screening 17.5 (2012): 629-40.
3) Savinainen, Juha R., Megumi Yoshino, Anna Minkkilä, Tapio Nevalainen, and Jarmo T. Laitinen. "Characterization of Binding Properties of Monoglyceride Lipase Inhibitors by a Versatile Fluorescence-based Technique." Analytical Biochemistry 399.1 (2010): 132-34
4) Taschler, U., F. P. W. Radner, C. Heier, R. Schreiber, M. Schweiger, G. Schoiswohl, K. Preiss-Landl, D. Jaeger, B. Reiter, H. C. Koefeler, J. Wojciechowski, C. Theussl, J. M. Penninger, A. Lass, G. Haemmerle, R. Zechner, and R. Zimmermann. "Monoglyceride Lipase Deficiency in Mice Impairs Lipolysis and Attenuates Diet-induced Insulin Resistance." Journal of Biological Chemistry 286.20 (2011): 17467-7477
External links
MGL
Inhibition
Endocannabinoids