This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox reserved 915
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
===Inhibition of MGL=== | ===Inhibition of MGL=== | ||
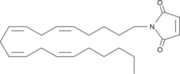
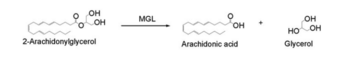
| - | The importance of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2013872/ Inhibition] of Monoglyceride lipase is to keep it from breaking down 2-arachidonoyl glycerol. When 2-Ag is broken down it is not able to suppress pain and depression brain functions that human beings experience. N-arachidonyl maleimide (NAM) is one inhibitor of MGL that reacts with the amino acid <scene name='58/580298/Cys252/1'>Cys252</scene>. '''Figure 2 | + | The importance of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2013872/ Inhibition] of Monoglyceride lipase is to keep it from breaking down 2-arachidonoyl glycerol. When 2-Ag is broken down it is not able to suppress pain and depression brain functions that human beings experience. N-arachidonyl maleimide (NAM) is one inhibitor of MGL that reacts with the amino acid <scene name='58/580298/Cys252/1'>Cys252</scene>. '''Figure 2''' |
[[Image:NAM.png|thumb|'''Figure 2:''' The structure of N-arachidonyl maleimide (NAM)that interacts with Cys252.]] | [[Image:NAM.png|thumb|'''Figure 2:''' The structure of N-arachidonyl maleimide (NAM)that interacts with Cys252.]] | ||
This Cysteine is buried in the active site near the catalytic serine and functions by sterically clashing with the natural ligand. A possible conformational change to Cys252 upon the binding of NAM could also lead to an inactive form of MGL. | This Cysteine is buried in the active site near the catalytic serine and functions by sterically clashing with the natural ligand. A possible conformational change to Cys252 upon the binding of NAM could also lead to an inactive form of MGL. | ||
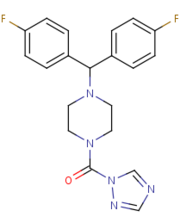
| - | MGL is also inhibited by being in complex with <scene name='58/580298/Sar629/2'>SAR629</scene> that is covalently bound to the catalytic Ser132. SAR629 adopts a Y shape and interacts with the MGL by hydrophobic interactions, with a few polar interactions as well. '''Figure 3 | + | MGL is also inhibited by being in complex with <scene name='58/580298/Sar629/2'>SAR629</scene> that is covalently bound to the catalytic Ser132. SAR629 adopts a Y shape and interacts with the MGL by hydrophobic interactions, with a few polar interactions as well. '''Figure 3''' |
[[Image:SAR.png|left|thumb|'''Figure 3:''' The structure and shape of SAR629.]] | [[Image:SAR.png|left|thumb|'''Figure 3:''' The structure and shape of SAR629.]] | ||
| Line 33: | Line 33: | ||
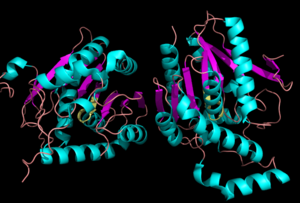
== Catalytic triad == | == Catalytic triad == | ||
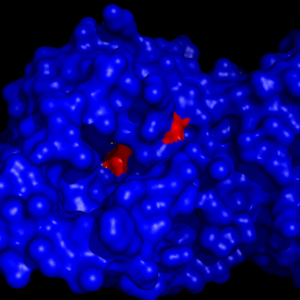
| - | MGL has a classic <scene name='58/580298/Catalytic_triad/1'>catalytic triad</scene> that contains Ser-His-Asp. The triad was found using site-directed mutagenesis of each individual residue and each of these amino acid residues are catalytically essential to MGL <ref name="Bertrand" />. The catalytic triad is located in the [[:Category:Ligand binding pocket| Binding Pocket]]buried at the bottom of it in the oxyanion hole connected by a water molecule.'''Figure 4 | + | MGL has a classic <scene name='58/580298/Catalytic_triad/1'>catalytic triad</scene> that contains Ser-His-Asp. The triad was found using site-directed mutagenesis of each individual residue and each of these amino acid residues are catalytically essential to MGL <ref name="Bertrand" />. The catalytic triad is located in the [[:Category:Ligand binding pocket| Binding Pocket]]buried at the bottom of it in the oxyanion hole connected by a water molecule.'''Figure 4''' |
[[Image:Catalytic_triad_binding_pocket.png|300px|thumb|'''Figure 4:''' The binding pocket of MGL with the catalytic triad (shown in red) buried in it.]] | [[Image:Catalytic_triad_binding_pocket.png|300px|thumb|'''Figure 4:''' The binding pocket of MGL with the catalytic triad (shown in red) buried in it.]] | ||
Revision as of 03:51, 9 April 2014
Monoglyceride Lipase (MGL)
| |||||||||||