We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 914
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
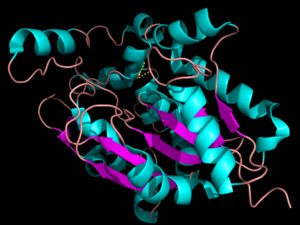
[[Image:pymol.png|300px|left|thumb|'''Figure 1:'''Three dimensional structure of Palmitoyl-Protein Thioesterase 1. The blue color represents the α-helices and the purple represents the β-sheets. The pink signifies random coil.]] | [[Image:pymol.png|300px|left|thumb|'''Figure 1:'''Three dimensional structure of Palmitoyl-Protein Thioesterase 1. The blue color represents the α-helices and the purple represents the β-sheets. The pink signifies random coil.]] | ||
| - | Palmitoyl-Protein Thioesterase 1 (PPT1) is a lysosomal enzyme that plays a role in the degradation of lipid-modified proteins<ref name="human">Palmitoyl-Protein Thioesterase 1 Precursor - Homo Sapiens. N.p., 1 Oct. 1996.</ref>. PPT1 | + | Palmitoyl-Protein Thioesterase 1 (PPT1) is a lysosomal enzyme that plays a role in the degradation of lipid-modified proteins<ref name="human">Palmitoyl-Protein Thioesterase 1 Precursor - Homo Sapiens. N.p., 1 Oct. 1996.</ref>. PPT1 derives its catalytic power from its catalytic triad, α/β hydrolase fold, and hydrophobic groove in order to remove fatty acid acyl groups, typically palmitate from cysteine residues in proteins<ref name="human"/>. PPT1 is able to be modified by cofactor enzymes, which can induce biological changes<ref name="human"/>. Misregulation of PPT1 modifications can cause various diseases, including infantile neuronal ceroid lipofuscinosis<ref name="PPT"/>, kufs disease<ref name="PPT"/>, and late-infantile neuronal ceroid lipofuscinosis<ref name="PPT"/>. Within these diseases, the production of PPT1 is decreased or eliminated completely, which leads to fatty acid buildup primarily in neuronal cells, leading to slowed developmental progress<ref name="PPT"/>. |
== Structure == | == Structure == | ||
| - | The secondary structure of PPT1 contains several α-helices and few β-sheets. PPT1 includes residues 28-306, after the 27-residue signal peptide has been removed <ref name="RSCB">Bellizzi III, John. RCSB Protein Data Bank. PNAS, 18 Apr. 2000.</ref>. | + | The secondary structure of PPT1 contains several α-helices and few β-sheets (Figure 1). PPT1 includes residues 28-306, after the 27-residue signal peptide has been removed <ref name="RSCB">Bellizzi III, John. RCSB Protein Data Bank. PNAS, 18 Apr. 2000.</ref>. An insertion is found between β6 and β7, residues 140-223, and that forms a <scene name='57/573128/9/1'>second domain</scene>, shown in blue, that is compromised almost entirely of the fatty acid binding site. This second domain region contains six helices, α2-α7<ref name="RSCB"/>. |
| - | === α/β Hydrolase Fold === | ||
The α/β Hydrolase Fold is common to many other hydrolases. The α/β hydrolase fold has a central 6 stranded parallel β-sheet consisting of <scene name='57/573128/4/1'>β3-β8</scene> and α-helices <scene name='57/573128/5/1'>αA, αB, αC, and αF</scene><ref name="RSCB"/>. It also consists of a catalytic triad and an oxyanion hole. The pKa of the nucleophile in the catalytic triad is lowered to allow the nucleophilic attack<ref name="Prom">Branneby, Cecilia. Exploiting Enzyme Promiscuity for Rational Design. KTH Biotechnology. N.p., May 2005</ref>. None of the enzymes within the α/β hydrolase fold family require a cofactor for catalytic activity. | The α/β Hydrolase Fold is common to many other hydrolases. The α/β hydrolase fold has a central 6 stranded parallel β-sheet consisting of <scene name='57/573128/4/1'>β3-β8</scene> and α-helices <scene name='57/573128/5/1'>αA, αB, αC, and αF</scene><ref name="RSCB"/>. It also consists of a catalytic triad and an oxyanion hole. The pKa of the nucleophile in the catalytic triad is lowered to allow the nucleophilic attack<ref name="Prom">Branneby, Cecilia. Exploiting Enzyme Promiscuity for Rational Design. KTH Biotechnology. N.p., May 2005</ref>. None of the enzymes within the α/β hydrolase fold family require a cofactor for catalytic activity. | ||
Revision as of 22:53, 9 April 2014
ββ
| This Sandbox is Reserved from Jan 06, 2014, through Aug 22, 2014 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 911 through Sandbox Reserved 922. |
To get started:
More help: Help:Editing |
Palmitoyl-Protein Thioesterase 1
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Palmitoyl-Protein Thioesterase 1 Precursor - Homo Sapiens. N.p., 1 Oct. 1996.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 PPT1. Genetics Home Reference. U.S. National Library of Medicine, Aug. 2015.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Bellizzi III, John. RCSB Protein Data Bank. PNAS, 18 Apr. 2000.
- ↑ 4.0 4.1 4.2 Branneby, Cecilia. Exploiting Enzyme Promiscuity for Rational Design. KTH Biotechnology. N.p., May 2005
External Resources
http://en.wikipedia.org/wiki/Palmitoyl_protein_thioesterase
https://www.counsyl.com/diseases/ppt1-related-neuronal-ceroid-lipofuscinosis/
http://en.wikipedia.org/wiki/Alpha/beta_hydrolase_fold
http://en.wikipedia.org/wiki/Catalytic_triad
http://www.biomedcentral.com/1471-2121/8/22
http://www.genecards.org/cgi-bin/carddisp.pl?gene=PPT1