We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 914
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
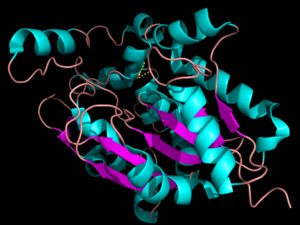
The secondary structure of PPT1 contains several α-helices and few β-sheets (Figure 1). PPT1 includes residues 28-306, after the 27-residue signal peptide has been removed <ref name="RSCB">Bellizzi III, John. RCSB Protein Data Bank. PNAS, 18 Apr. 2000.</ref>. An insertion is found between β6 and β7, residues 140-223, and that forms a <scene name='57/573128/9/1'>second domain</scene>, shown in blue, that is compromised almost entirely of the fatty acid binding site. This second domain region contains six helices, α2-α7<ref name="RSCB"/>. | The secondary structure of PPT1 contains several α-helices and few β-sheets (Figure 1). PPT1 includes residues 28-306, after the 27-residue signal peptide has been removed <ref name="RSCB">Bellizzi III, John. RCSB Protein Data Bank. PNAS, 18 Apr. 2000.</ref>. An insertion is found between β6 and β7, residues 140-223, and that forms a <scene name='57/573128/9/1'>second domain</scene>, shown in blue, that is compromised almost entirely of the fatty acid binding site. This second domain region contains six helices, α2-α7<ref name="RSCB"/>. | ||
| - | The α/β | + | The α/β hydrolase fold is common to many other hydrolases <ref name="RSCB"/>. The α/β hydrolase fold has a central 6 stranded parallel β-sheet consisting of <scene name='57/573128/4/1'>β3-β8</scene> and α-helices <scene name='57/573128/5/1'>αA, αB, αC, and αF</scene><ref name="RSCB"/>. A catalytic triad and an oxyanion hole are also common features to the PPT1 protein family. None of the enzymes within the α/β hydrolase fold family require a cofactor for catalytic activity. |
=== Catalytic Triad === | === Catalytic Triad === | ||
| - | The <scene name='57/573128/2/1'>catalytic triad</scene> is composed of Ser115, His289, and Asp233, which is the same as the catalytic triad in chymotrypsin. | + | The <scene name='57/573128/2/1'>catalytic triad</scene> is composed of Ser115, His289, and Asp233, which is the same as the catalytic triad in chymotrypsin <ref name="human"/>. A water molecule is occupying the <scene name='57/573128/7/1'>oxyanion hole</scene> and it is hydrogen bonded to Ser115 <ref name="Prom"/>. The purpose of the oxyanion hole is to stabilize the oxyanion that is formed after the nucleophilic attack of the transition state. Ser115 acts as a nucleophile, while His289 and Asp233 are coordinated to Ser115 to lower its pKa value so it can undergo catalytic activity<ref name="Prom"/>. The pKa of the nucleophile in the catalytic triad is lowered to allow the nucleophilic attack<ref name="Prom">Branneby, Cecilia. Exploiting Enzyme Promiscuity for Rational Design. KTH Biotechnology. N.p., May 2005</ref>. |
| - | A water molecule is occupying the <scene name='57/573128/7/1'>oxyanion hole</scene> and it is hydrogen bonded to Ser115 <ref name="Prom"/>. The purpose of the oxyanion hole is to stabilize the oxyanion that is formed after the nucleophilic attack | + | |
===Hydrophobic Groove === | ===Hydrophobic Groove === | ||
| - | The <scene name='57/573128/3/1'>hydrophobic binding groove</scene> is located in the second domain of PPT1, where palmitate mainly binds. The fact that palmitate has to <scene name='57/573128/6/1'>bend</scene> to fit into the binding pocket suggests that this pocket is designed to bind an unsaturated fatty acid, with a possible cis-double bond between C4 and C5<ref name="RSCB"/>. The top portion of the groove is formed by the residues from α2 to α3. Several residues that are present near the active site create the rest of the groove, including Ile235, Val236, Gln116, Gly40, and Met41<ref name="RSCB"/>. | + | The <scene name='57/573128/3/1'>hydrophobic binding groove</scene> is located in the second domain of PPT1, where palmitate mainly binds. The fact that palmitate has to <scene name='57/573128/6/1'>bend</scene> to fit into the binding pocket suggests that this pocket is designed to bind an unsaturated fatty acid, with a possible cis-double bond between C4 and C5<ref name="RSCB"/>. The top portion of the groove is formed by the residues from α2 to α3. Several residues that are present near the active site create the rest of the groove, including <scene name='57/573128/10/1'>Ile235, Val236, Gln116, Gly40, and Met41</scene><ref name="RSCB"/>. |
== Function == | == Function == | ||
Revision as of 23:10, 9 April 2014
ββ
| This Sandbox is Reserved from Jan 06, 2014, through Aug 22, 2014 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 911 through Sandbox Reserved 922. |
To get started:
More help: Help:Editing |
Palmitoyl-Protein Thioesterase 1
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Palmitoyl-Protein Thioesterase 1 Precursor - Homo Sapiens. N.p., 1 Oct. 1996.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 PPT1. Genetics Home Reference. U.S. National Library of Medicine, Aug. 2015.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 Bellizzi III, John. RCSB Protein Data Bank. PNAS, 18 Apr. 2000.
- ↑ 4.0 4.1 4.2 Branneby, Cecilia. Exploiting Enzyme Promiscuity for Rational Design. KTH Biotechnology. N.p., May 2005
External Resources
http://en.wikipedia.org/wiki/Palmitoyl_protein_thioesterase
https://www.counsyl.com/diseases/ppt1-related-neuronal-ceroid-lipofuscinosis/
http://en.wikipedia.org/wiki/Alpha/beta_hydrolase_fold
http://en.wikipedia.org/wiki/Catalytic_triad
http://www.biomedcentral.com/1471-2121/8/22
http://www.genecards.org/cgi-bin/carddisp.pl?gene=PPT1