Sandbox reserved 919
From Proteopedia
| Line 11: | Line 11: | ||
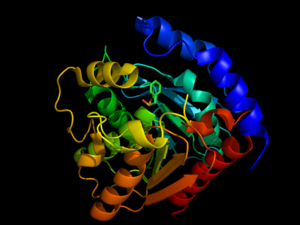
<StructureSection load='3DNM' size='350' frame='true' align='right' caption='Hormone-Sensitive Lipase from [[3dnm]]' scene='58/580297/3dnm_cartoon/2' > | <StructureSection load='3DNM' size='350' frame='true' align='right' caption='Hormone-Sensitive Lipase from [[3dnm]]' scene='58/580297/3dnm_cartoon/2' > | ||
| - | <scene name='58/580297/3dnm_cartoon_dotsribbon/1'>Hormone-sensitive lipases</scene> are generally well-conserved across domains, including prokaryotes, showing 29, 26, and 22% residue overlap in [http://en.wikipedia.org/wiki/Alicyclobacillus ''Alicyclobacillus acidocaldarius''], [http://en.wikipedia.org/wiki/Archaeoglobus ''Archaeoglobus fulgidus''], and [http://en.wikipedia.org/wiki/Bacillus_subtilis ''Bacillus subtilis''], respectively. <ref name="Nam">PMID:19089974</ref> HSL is composed of two main structural domains, consisting of a slightly variable N-terminus that is thought to contribute to numerous factors including activity, specificity, regioselectivity, thermophilicity, and thermostability. <ref name="Nam">PMID:19089974</ref> The second, highly conserved, domain of HSL is the C-terminal catalytic domain, which contains the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad], viewed <scene name='58/580297/3dnm_triad_zoomedout/1'>here</scene>, a charge relay network that is characteristic of many hydrolases. Size-exclusion chromatography studies have shown that HSL has a ligand pocket that is approximately 16Å deep, suggesting that HSL primarily hydrolyzes shorter chained molecules. <ref name="Nam">PMID:19089974</ref> | + | <scene name='58/580297/3dnm_cartoon_dotsribbon/1'>Hormone-sensitive lipases</scene> are generally well-conserved across domains, including prokaryotes, showing 29, 26, and 22% residue overlap in [http://en.wikipedia.org/wiki/Alicyclobacillus ''Alicyclobacillus acidocaldarius''], [http://en.wikipedia.org/wiki/Archaeoglobus ''Archaeoglobus fulgidus''], and [http://en.wikipedia.org/wiki/Bacillus_subtilis ''Bacillus subtilis''], respectively. <ref name="Nam">PMID:19089974</ref> HSL is composed of two main structural domains, consisting of a slightly variable N-terminus (shown in blue in the <scene name='58/580297/3dnm_cartoon/3'>default view</scene>) that is thought to contribute to numerous factors including activity, specificity, regioselectivity, thermophilicity, and thermostability. <ref name="Nam">PMID:19089974</ref> The second, highly conserved, domain of HSL is the C-terminal catalytic domain (colors other than blue), which contains the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad], viewed <scene name='58/580297/3dnm_triad_zoomedout/1'>here</scene>, a charge relay network that is characteristic of many hydrolases. Size-exclusion chromatography studies have shown that HSL has a ligand pocket that is approximately 16Å deep, suggesting that HSL primarily hydrolyzes shorter chained molecules. <ref name="Nam">PMID:19089974</ref> |
The catalytic triad <scene name='58/580297/3dnm_triad_zoomedin/1'>situates itself</scene> toward the middle of HSL. The catalytic triad is composed of residues <scene name='58/580297/3dnm_ligandsite_triad_chains/4'>Ser157, Glu251, and His281</scene>. The Ser157 residue sits at a site deemed the "nucleophilic elbow," that models an approximate torsion of Φ = 60° and Ψ =-120°. This nucleophilic elbow is stabilized by a hydrogen bond between the proximal nitrogen and oxygen atoms of His281 and Glu251, respectively. This model also shows the strong nucleophilic character of Ser157, portraying the covalent binding to <scene name='58/580297/3dnm_ligandsite_triad_chains/3'>β-mercaptoethanol</scene>. Return to default view, <scene name='58/580297/3dnm_cartoon/3'>here</scene>. | The catalytic triad <scene name='58/580297/3dnm_triad_zoomedin/1'>situates itself</scene> toward the middle of HSL. The catalytic triad is composed of residues <scene name='58/580297/3dnm_ligandsite_triad_chains/4'>Ser157, Glu251, and His281</scene>. The Ser157 residue sits at a site deemed the "nucleophilic elbow," that models an approximate torsion of Φ = 60° and Ψ =-120°. This nucleophilic elbow is stabilized by a hydrogen bond between the proximal nitrogen and oxygen atoms of His281 and Glu251, respectively. This model also shows the strong nucleophilic character of Ser157, portraying the covalent binding to <scene name='58/580297/3dnm_ligandsite_triad_chains/3'>β-mercaptoethanol</scene>. Return to default view, <scene name='58/580297/3dnm_cartoon/3'>here</scene>. | ||
Revision as of 17:08, 19 April 2014
Contents |
Introduction to hormone-sensitive lipase
Hormone-sensitive lipases (HSL) represent a class of esterases within the α/β hydrolase family. HSL catalyzes the cleavage of ester bonds in fatty acid molecules when stimulated by a hormone. [1] The activation and mobilization of hormone-sensitive lipase can be triggered by various catecholamines and inhibited by insulin. [2] Catecholamines, such as epinephrine, are rapidly spread throughout the body during times of energy mobilization, like in the fight of flight response. Conversely, insulin triggers glucose uptake, requiring the storage of energy, opposing HSL's function. HSL is clinically relevant, because the mobilization of or inability to mobilize fats in cells is directly related to fat accumulation seen in artherosclerosisand obesity.[3] Such diseases are characterized by an accumulation of fats and researchers are investigating whether HSL's activity plays a role or not. [2] Investigation of HSL's structure and function could provide a better clinical understanding of these diseases. [3]
Briefly, HSL is stimulated by the binding of catecholamines to β-adrenergic receptors. β-adrenergic receptors coupled with adenylate cyclase (AC) then stimulate G-proteins to increase the levels of cystolic cAMP. Elevated levels of cAMP leads to an activation protein kinase A (PKA) leading to phosphorylation of serine residues on HSL activating and translocating HSL to lipid droplets for lipolysis. Conversely, insulin signaling decreases cystolic cAMP levels, resulting in a decreased HSL mobilization. [1]
Structure of hormone-sensitive lipase
| |||||||||||
Additional pages about hormone-sensitive lipase
References
- ↑ 1.0 1.1 Holm C. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem Soc Trans. 2003 Dec;31(Pt 6):1120-4. PMID:14641008 doi:http://dx.doi.org/10.1042/
- ↑ 2.0 2.1 Ray H, Beylot M, Arner P, Larrouy D, Langin D, Holm C, Large V. The presence of a catalytically inactive form of hormone-sensitive lipase is associated with decreased lipolysis in abdominal subcutaneous adipose tissue of obese subjects. Diabetes. 2003 Jun;52(6):1417-22. PMID:12765952
- ↑ 3.0 3.1 Yeaman SJ. Hormone-sensitive lipase--new roles for an old enzyme. Biochem J. 2004 Apr 1;379(Pt 1):11-22. PMID:14725507 doi:http://dx.doi.org/10.1042/BJ20031811
- ↑ 4.0 4.1 4.2 Nam KH, Kim MY, Kim SJ, Priyadarshi A, Kwon ST, Koo BS, Yoon SH, Hwang KY. Structural and functional analysis of a novel hormone-sensitive lipase from a metagenome library. Proteins. 2009 Mar;74(4):1036-40. PMID:19089974 doi:http://dx.doi.org/10.1002/prot.22313
- ↑ Kanwar SS, Kaushal RK, Jawed A, Gupta R, Chimni SS. Methods for inhibition of residual lipase activity in colorimetric assay: a comparative study. Indian J Biochem Biophys. 2005 Aug;42(4):233-7. PMID:23923547
- ↑ Kraemer FB, Shen WJ. Hormone-sensitive lipase: control of intracellular tri-(di-)acylglycerol and cholesteryl ester hydrolysis. J Lipid Res. 2002 Oct;43(10):1585-94. PMID:12364542