User:Brittany Carroll/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Introduction == | == Introduction == | ||
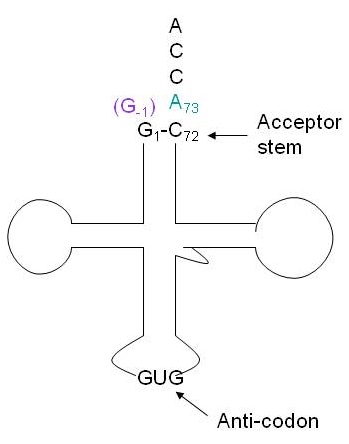
| - | + | [[Image:trnahis.jpg]] | |
| - | + | ||
== Classification == | == Classification == | ||
Revision as of 00:58, 24 April 2014
tRNA(His) guanylyltransferase
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644