We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 191
From Proteopedia
(Difference between revisions)
| Line 35: | Line 35: | ||
===Mutations leading to Infantile Neuronal Ceroid Lipofuscinosis=== | ===Mutations leading to Infantile Neuronal Ceroid Lipofuscinosis=== | ||
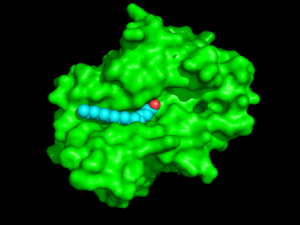
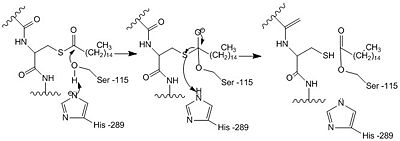
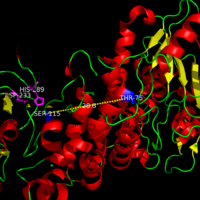
| - | Various mutations have been found in INCL patients<ref name="Ryan-1">PMID:10191107</ref>. Most of these mutations are caused by nonsense or missense mutations within close proximity to the catalytic <scene name='43/436866/Triad_w_zoom_no_backbones/1'>triad</scene>. These mutations lead to an inactive PPT-1 enzyme as they are predicted to create unfavorable steric, polar, and electrostatic interactions that could disturb the nucleophilic elbow <ref name="mutations" />. The nucleophilic elbow is responsible for proper location and orientation of the Ser-115. The catalytic activity of PPT-1 is greatly reduced if the positioning of Ser-115 is altered, as Ser-115 must be properly orientated to be activated by His-289 to be positioned to attack the substrate. An example of a INCL mutation such as <scene name='58/580837/Methionine/9'>Val181Met</scene> and <scene name='58/580837/Lysine_mutation/4'>Glu184Lys</scene> gives a good depiction of how the increase in size in the mutated amino acids and positive charge on the inserted lysine residue would create steric and polar clashes with the adjacent helices of the binding pocket compared to the <scene name='58/580837/Val181glu184/3'>normal Val-181 and Glu-184</scene>. <scene name='58/580837/Arginine_fine/5'> | + | Various mutations have been found in INCL patients<ref name="Ryan-1">PMID:10191107</ref>. Most of these mutations are caused by nonsense or missense mutations within close proximity to the catalytic <scene name='43/436866/Triad_w_zoom_no_backbones/1'>triad</scene>. These mutations lead to an inactive PPT-1 enzyme as they are predicted to create unfavorable steric, polar, and electrostatic interactions that could disturb the nucleophilic elbow <ref name="mutations" />. The nucleophilic elbow is responsible for proper location and orientation of the Ser-115. The catalytic activity of PPT-1 is greatly reduced if the positioning of Ser-115 is altered, as Ser-115 must be properly orientated to be activated by His-289 to be positioned to attack the substrate. An example of a INCL mutation such as <scene name='58/580837/Methionine/9'>Val181Met</scene> and <scene name='58/580837/Lysine_mutation/4'>Glu184Lys</scene> gives a good depiction of how the increase in size in the mutated amino acids and positive charge on the inserted lysine residue would create steric and polar clashes with the adjacent helices of the binding pocket compared to the <scene name='58/580837/Val181glu184/3'>normal Val-181 and Glu-184</scene>. <scene name='58/580837/Arginine_fine/5'>Normal Arg-122</scene> which is below <ref name="Ryan-1">PMID:10191107</ref>. |
(*All mutation scenes are the proposed best rotamers of the mutation. These mutations were created through the use of the mutagenesis wizard in the PyMOL program) | (*All mutation scenes are the proposed best rotamers of the mutation. These mutations were created through the use of the mutagenesis wizard in the PyMOL program) | ||
Revision as of 16:56, 25 April 2014
| This Sandbox is Reserved from Feb 02, 2011, through Jul 31, 2011 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 191 through Sandbox Reserved 200. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Wang R, Borazjani A, Matthews AT, Mangum LC, Edelmann MJ, Ross MK. Identification of palmitoyl protein thioesterase 1 in human THP1 monocytes and macrophages and characterization of unique biochemical activities for this enzyme. Biochemistry. 2013 Oct 29;52(43):7559-74. doi: 10.1021/bi401138s. Epub 2013 Oct, 18. PMID:24083319 doi:http://dx.doi.org/10.1021/bi401138s
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Bellizzi JJ 3rd, Widom J, Kemp C, Lu JY, Das AK, Hofmann SL, Clardy J. The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis. Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4573-8. PMID:10781062 doi:10.1073/pnas.080508097
- ↑ 3.0 3.1 Simonati A, Tessa A, Bernardina BD, Biancheri R, Veneselli E, Tozzi G, Bonsignore M, Grosso S, Piemonte F, Santorelli FM. Variant late infantile neuronal ceroid lipofuscinosis because of CLN1 mutations. Pediatr Neurol. 2009 Apr;40(4):271-6. doi: 10.1016/j.pediatrneurol.2008.10.018. PMID:19302939 doi:http://dx.doi.org/10.1016/j.pediatrneurol.2008.10.018
- ↑ 4.0 4.1 Hofmann SL, Das AK, Yi W, Lu JY, Wisniewski KE. Genotype-phenotype correlations in neuronal ceroid lipofuscinosis due to palmitoyl-protein thioesterase deficiency. Mol Genet Metab. 1999 Apr;66(4):234-9. PMID:10191107 doi:http://dx.doi.org/10.1006/mgme.1999.2803
- ↑ 5.0 5.1 Dawson G, Schroeder C, Dawson PE. Palmitoyl:protein thioesterase (PPT1) inhibitors can act as pharmacological chaperones in infantile Batten disease. Biochem Biophys Res Commun. 2010 Apr 23;395(1):66-9. doi:, 10.1016/j.bbrc.2010.03.137. Epub 2010 Mar 25. PMID:20346914 doi:http://dx.doi.org/10.1016/j.bbrc.2010.03.137
- ↑ Vines DJ, Warburton MJ. Classical late infantile neuronal ceroid lipofuscinosis fibroblasts are deficient in lysosomal tripeptidyl peptidase I. FEBS Lett. 1999 Jan 25;443(2):131-5. PMID:9989590
- ↑ Mitchison HM, Taschner PE, Kremmidiotis G, Callen DF, Doggett NA, Lerner TJ, Janes RB, Wallace BA, Munroe PB, O'Rawe AM, Gardiner RM, Mole SE. Structure of the CLN3 gene and predicted structure, location and function of CLN3 protein. Neuropediatrics. 1997 Feb;28(1):12-4. PMID:9151311 doi:http://dx.doi.org/10.1055/s-2007-973656
External Resources
Gauche Effect Wikipedia page
Palmitic acid Wikipedia page
Infantile Neuronal Ceroid Lipofuscinosis Wikipedia page
PMSF Wikipedia page
Protein Chaperones Wikipedia page
Endoplasmic reticulum Wikipedia page
Palmitoylation Wikipedia page