You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
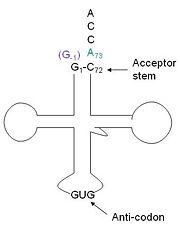
tRNA His, highlighting the incoming G-1 (purple) opposite A73 (green)
tRNA His (link to wikipedia) has a guanine monophosphate (GMP) residue at the 5’ end in all domains of life, besides α-proteopbacteria. This GMP is referred to as G-1. In prokaryotes (link to wikipedia) G-1 is encoded in the genome. RNase P (link to wikipedia) cleaves pre-tRNAHis to generate the mature tRNA, leaving an extra basepair on the acceptor stem, G-1:C73. In eukaryotes the G-1 residue is not encoded and needs to be added post-transcription. The enzyme that catalyzes this reaction is the polymerase, tRNAHis guanylyltransferase (Thg1). Howerver, the addition of the GMP residue is nontemplated, inserting GMP across from A73 in the acceptor stem creating a mismatch. Unlike most polymerases, Thg1 adds nucleotides in the 3’ –to- 5’ direction, while forming a normal 3’ –to- 5’ phosphodiester bond. Therefore, the 3’-OH of the incoming nucleotide attacks the 5’ end of the polynucleotide chain. This is a two step mechanism where the polynucleotide chain is first adenylated and then guanylated.
This addition is interesting for multiple reasons. It is one of only a few known reactions where a normal 3’-to-5’ phosphodiester bond is formed in a 3’ –to- 5’ direction. Also, the additional 5’ nucleotide is unique to tRNAHis, with the exception of a tRNAPhe species. Lastly, this modification is essential, at least in yeast.
Classification
CATH
Class: Mixed α & β
Architecture: 2-layer Sandwich
Topology: Alpha-Beta Plaits
Homologous Superfamily: Adenylyl Cyclase
3.30.70.1230
SCOP
Class: Alpha and Beta proteins
Fold: Ferredoxin-like
Superfamily: Nucleotide cyclase
Family: Adenylyl and guanylyl cylclase catalytic domain
Structure
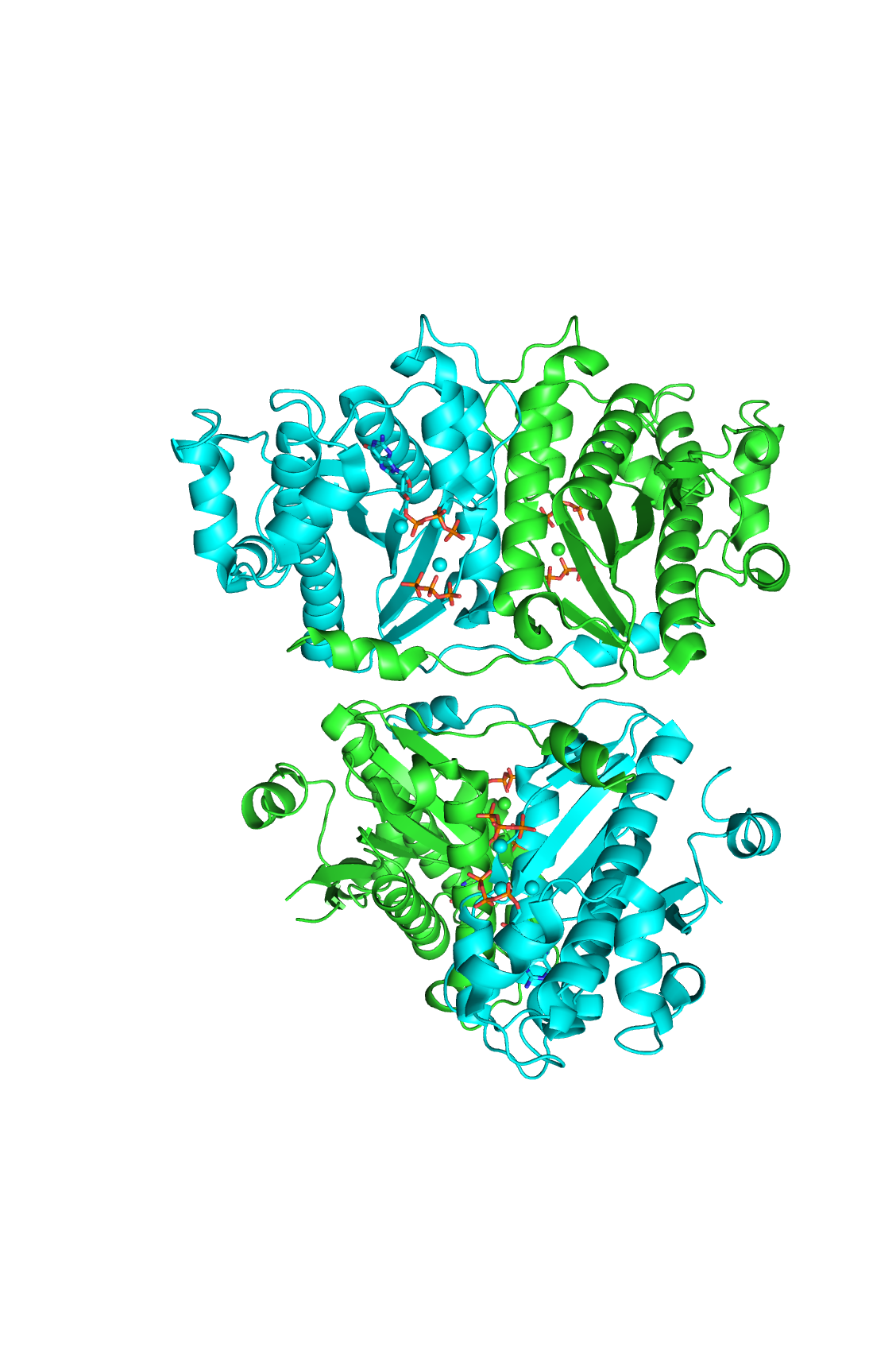
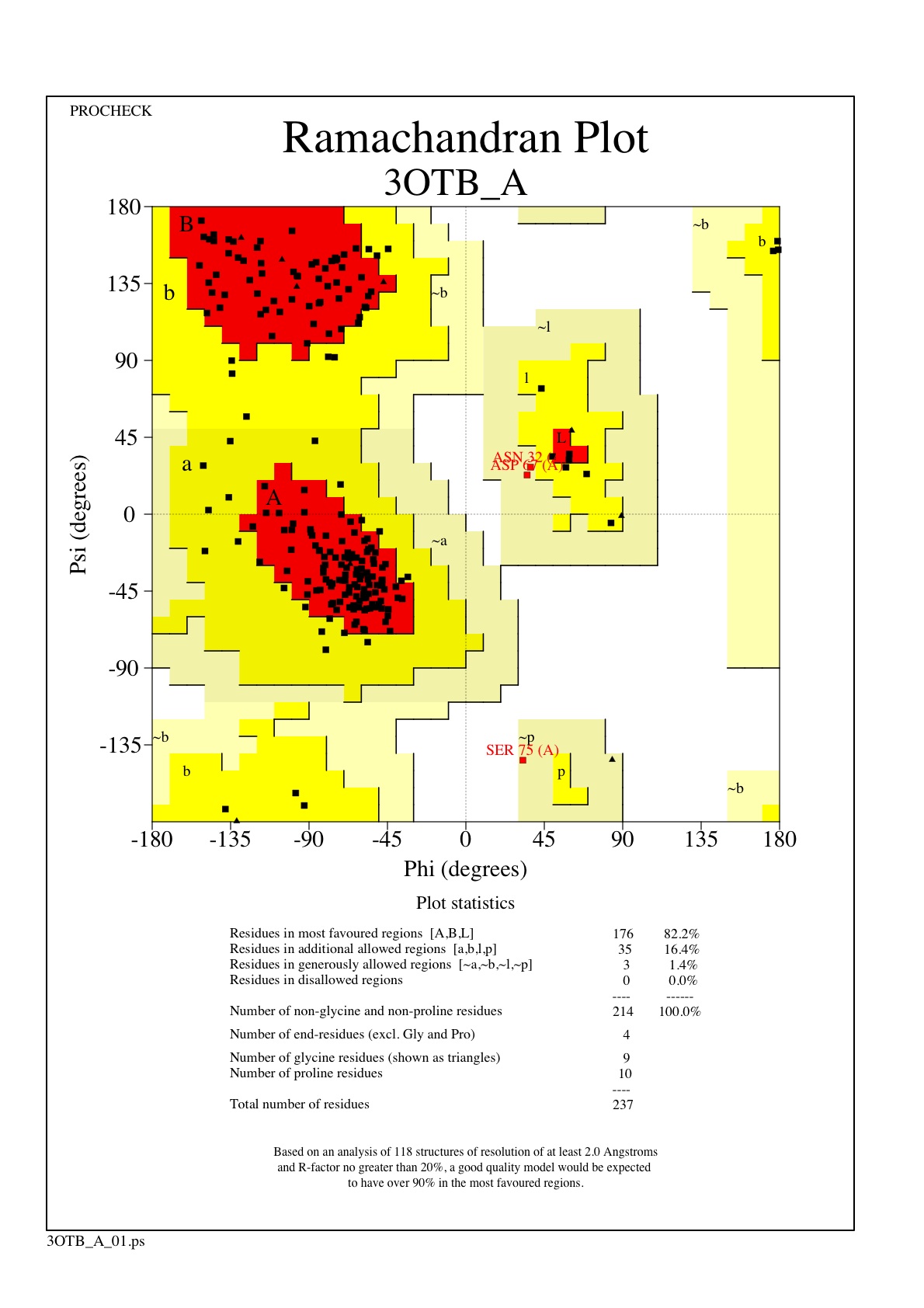
Structural highlights
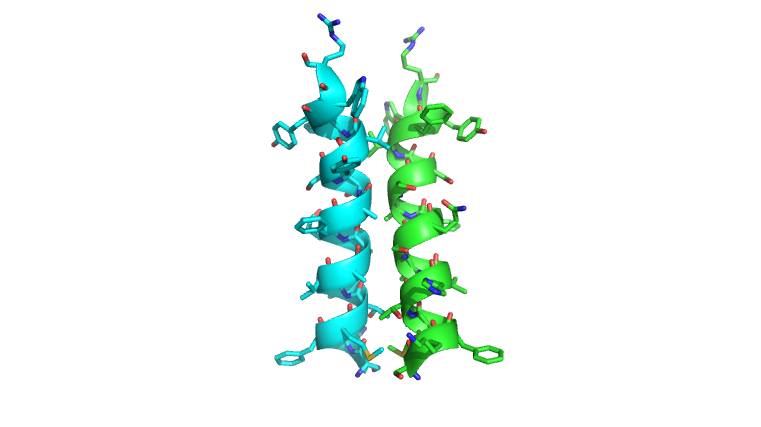
N-terminal helix cap
The N-terminal cap of the helix follows a Ib motiff. This motiff is also known as a capping box.
N’ -> N4 h-xpxph
N’- P S N Q T L –N4 (residues 135-140 in 3otb)
Hydrogen bonds between N’ and the backbone of N3 and N3 with N’ backbone are shown in the figure. The figure is difficult to see the T with P bb but it is not linear, this may just be due to modeling as it is close enough to form a h-bond. There is also a hydrophobic interaction between P and L.
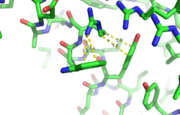
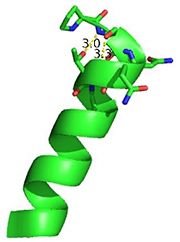
This image depicts two cation-π interactions between Arg and Tyr or Trp. The energetic significances are -1.22 and -6.55 kj/mol respectively. (site website) (30tb)
A cation-π interaction occurs between a cation and the face of a simple aromatic, there is partial negative charge in the center of the ring. The cation-π interaction is actually stronger than a salt bridge because of the desolvation penalty. With the cation-π interaction the cation has a similar dosolvation penalty to pay as the salt bridge ions but the π system is already poorly solvated. Also there is not neutralization of charge that occurs between the two groups. These properties of the cation-π interaction imply that thecation-π interactions on protein surfaces (mainly where they are seen) could contribute to protein structure and stability.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.