User:Brittany Carroll/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
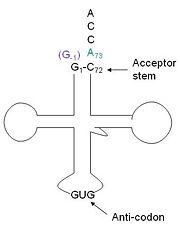
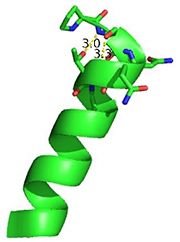
[[Image:Trnahis.jpg|left|thumb|tRNA His, highlighting the incoming G-1 (purple) opposite A73 (green)]] | [[Image:Trnahis.jpg|left|thumb|tRNA His, highlighting the incoming G-1 (purple) opposite A73 (green)]] | ||
| - | tRNA His | + | [http://www.http://en.wikipedia.org/wiki/Transfer_RNA tRNA]His has a [http://www.http://en.wikipedia.org/wiki/Guanosine_monophosphate guanine monophosphate] (GMP) residue at the 5’ end in all domains of life, besides α-proteopbacteria. This GMP is referred to as G-1. In prokaryotes (link to wikipedia) G-1 is encoded in the genome. RNase P (link to wikipedia) cleaves pre-tRNAHis to generate the mature tRNA, leaving an extra basepair on the acceptor stem, G-1:C73. In eukaryotes the G-1 residue is not encoded and needs to be added post-transcription. The enzyme that catalyzes this reaction is the polymerase,[http://www.en.wikipedia.org/wiki/Guanylyl_transferase tRNAHis guanylyltransferase] (Thg1). Howerver, the addition of the GMP residue is nontemplated, inserting GMP across from A73 in the acceptor stem creating a mismatch. Unlike most polymerases, Thg1 adds nucleotides in the 3’ –to- 5’ direction, while forming a normal 3’ –to- 5’ phosphodiester bond. Therefore, the 3’-OH of the incoming nucleotide attacks the 5’ end of the polynucleotide chain. This is a two step mechanism where the polynucleotide chain is first adenylated and then guanylated. |
This addition is interesting for multiple reasons. It is one of only a few known reactions where a normal 3’-to-5’ phosphodiester bond is formed in a 3’ –to- 5’ direction. Also, the additional 5’ nucleotide is unique to tRNAHis, with the exception of a tRNAPhe species. Lastly, this modification is essential, at least in yeast. | This addition is interesting for multiple reasons. It is one of only a few known reactions where a normal 3’-to-5’ phosphodiester bond is formed in a 3’ –to- 5’ direction. Also, the additional 5’ nucleotide is unique to tRNAHis, with the exception of a tRNAPhe species. Lastly, this modification is essential, at least in yeast. | ||
Revision as of 02:04, 27 April 2014
tRNA(His) guanylyltransferase
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644