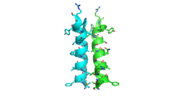We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Brittany Carroll/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
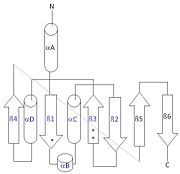
[[Image:thg1topology.jpg|right|thumb| This 2D topology diagram shows the βαββαβ fold of Thg1. The helices and strands involved in the fold are in blue font. The fold is most similar to that of cylcases. The mechanism is more likely the same as family A polymerases, with the conserved carboxylates shown as asterisks(*). [[3otb]]]] | [[Image:thg1topology.jpg|right|thumb| This 2D topology diagram shows the βαββαβ fold of Thg1. The helices and strands involved in the fold are in blue font. The fold is most similar to that of cylcases. The mechanism is more likely the same as family A polymerases, with the conserved carboxylates shown as asterisks(*). [[3otb]]]] | ||
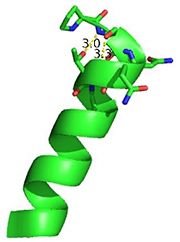
| - | Interestingly, Thg1 shares structural similarities to both [http://www.en.wikipedia.org/wiki/Cyclase cyclases] and the palm domain of canonical [http://www.en.wikipedia.org/wiki/DNA_polymerase polymerases], without sequence similarities. The βαββαβ motif is most homologous with adenylyl and guanylyl cyclases. However, based upon the model the mechanism seems to be more like that of a family A polymerase. The model suggests Thg1 has three catalytic carboxylates: aspartate 29, aspartate 76, and glutamate 77. Cyclases only have two catalytic carboxylates, two aspartates and either cysteine, alanine, or glycine. The position of the carboxylates in Thg1 is homologous to those of [http://www.en.wikipedia.org/wiki/T7_DNA_polymerase T7 DNA Polymerase]. An overlay of the palm domain of T7 and Thg1 shows that the three carboxylates, two metal ions, and the incoming nucleotide are conserved and in similar postions. This indicates that Thg1 most likely uses the two-metal-ion mechaism of canonical 5' to 3' polymerases.<ref>PMID:21059936</ref> | + | Interestingly, Thg1 shares structural similarities to both [http://www.en.wikipedia.org/wiki/Cyclase cyclases] and the palm domain of canonical [http://www.en.wikipedia.org/wiki/DNA_polymerase polymerases], without sequence similarities. The βαββαβ motif is most homologous with adenylyl and guanylyl cyclases. However, based upon the model the mechanism seems to be more like that of a family A polymerase. The model suggests Thg1 has three catalytic <scene name='58/582908/Active_site/1'>carboxylates</scene>: aspartate 29, aspartate 76, and glutamate 77. Cyclases only have two catalytic carboxylates, two aspartates and either cysteine, alanine, or glycine. The position of the carboxylates in Thg1 is homologous to those of [http://www.en.wikipedia.org/wiki/T7_DNA_polymerase T7 DNA Polymerase]. An overlay of the palm domain of T7 and Thg1 shows that the three carboxylates, two metal ions, and the incoming nucleotide are conserved and in similar postions. This indicates that Thg1 most likely uses the two-metal-ion mechaism of canonical 5' to 3' polymerases.<ref>PMID:21059936</ref> |
== Structure == | == Structure == | ||
| Line 20: | Line 20: | ||
[[Image:alpha_dimer.png|300|right|thumb|The interface between alpha helices D of the two monomers shows the large amount of contacts helping to stablize the dimer.[[3otb]]]] | [[Image:alpha_dimer.png|300|right|thumb|The interface between alpha helices D of the two monomers shows the large amount of contacts helping to stablize the dimer.[[3otb]]]] | ||
| - | Each monomer has an antiparallel β-sheet with six strands and seven α-helices around the sheet. Thg1 forms a homotetramer, with extensive contacts between the dimer. Even though there are fewer contacts between the dimer of dimers, the tetramer is the most stable oligomeric form. | + | Each monomer has an antiparallel β-sheet with six strands and seven α-helices around the sheet. Thg1 forms a homotetramer, with extensive contacts between the <scene name='58/582908/Space_fill_dimer/1'>dimer</scene>. Even though there are fewer contacts between the dimer of dimers, the tetramer is the most stable oligomeric form. |
The dimer is stabilized mainly by hydrogen bonds from αD and β4. There are also two salt bridges that help hold the dimer together: Lys to Asp and Glu to Arg (chain A to B). <ref>PMID:21059936</ref> | The dimer is stabilized mainly by hydrogen bonds from αD and β4. There are also two salt bridges that help hold the dimer together: Lys to Asp and Glu to Arg (chain A to B). <ref>PMID:21059936</ref> | ||
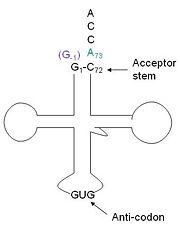
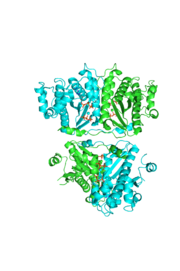
| - | The reason for the tetramer remained elusive until the "Candida albicans" Thg1 with tRNA His was solved. The model showed that two tRNA His molecules are coordinated between the four subunits, each interacting with three monomers. The acceptor stem of the tRNA is coordinated by the dimer of monomers, and the anitcodon is recognized by the second dimer. The binding to the third subunit, is necessary for recognition but may also aid in the correct positioning for guanylylation. Thg1 is selective to binding tRNA His through the recognition of the anticodon loop with base stacking and hydrogen bonding. G34 is recognized via aromatic stacking, between Phe194 and G37(tRNA), this interaction is not base specific but it is purine specific. Asn202 coordinates U35 through hydrogen bonding and His154 base stacks with the third base G36. These interactions cause distortion of the loop that is stabilized by hydrogen bonds from other crucial surrounding amino acids. The structure with tRNA also lends insight into why Thg1 works in is reverse. It appears that Thg1 is a mirror image of forward polymerases, suggesting that the direction in which the template approaches the active site is important in the direction of addition.<ref>PMID:24324136</ref> | + | The reason for the tetramer remained elusive until the "Candida albicans" Thg1 with tRNA His was solved. The model showed that two tRNA His molecules are coordinated between the <scene name='58/582908/Tetramer_rna/1'>four subunits</scene>, each interacting with three monomers. The acceptor stem of the tRNA is coordinated by the dimer of monomers, and the anitcodon is recognized by the second dimer. The binding to the third subunit, is necessary for recognition but may also aid in the correct positioning for guanylylation. Thg1 is selective to binding tRNA His through the recognition of the anticodon loop with base stacking and hydrogen bonding. G34 is recognized via aromatic stacking, between Phe194 and G37(tRNA), this interaction is not base specific but it is purine specific. Asn202 coordinates U35 through hydrogen bonding and His154 base stacks with the third base G36. These interactions cause distortion of the loop that is stabilized by hydrogen bonds from other crucial surrounding amino acids. The structure with tRNA also lends insight into why Thg1 works in is reverse. It appears that Thg1 is a mirror image of forward polymerases, suggesting that the direction in which the template approaches the active site is important in the direction of addition.<ref>PMID:24324136</ref> |
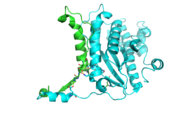
[[Image:tetra.png|300|left|thumb|The hThg1 tetramer is shown in cartoon form. The dimer of dimers is crystallographic. The dGTP and triphosphate are shown in sticks, with the coordination metal ions as balls.[[3otb]]]] | [[Image:tetra.png|300|left|thumb|The hThg1 tetramer is shown in cartoon form. The dimer of dimers is crystallographic. The dGTP and triphosphate are shown in sticks, with the coordination metal ions as balls.[[3otb]]]] | ||
Revision as of 01:12, 28 April 2014
tRNA(His) guanylyltransferase
| |||||||||||
Addition SD Structures of Thg1
3otc, 3otd, 3ote - Thg1 - Homo sapiens
4kgk, 4kgm - Thg1-like - Bacillus thuringiensis
3wbz, 3wc0, 3wc1, 3wc2 - Thg1 - Candida albicans
References
- ↑ Jackman JE, Gott JM, Gray MW. Doing it in reverse: 3'-to-5' polymerization by the Thg1 superfamily. RNA. 2012 May;18(5):886-99. doi: 10.1261/rna.032300.112. Epub 2012 Mar 28. PMID:22456265 doi:http://dx.doi.org/10.1261/rna.032300.112
- ↑ Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases. Proc Natl Acad Sci U S A. 2010 Nov 8. PMID:21059936 doi:10.1073/pnas.1010436107
- ↑ Hyde SJ, Rao BS, Eckenroth BE, Jackman JE, Doublie S. Structural Studies of a Bacterial tRNA(HIS) Guanylyltransferase (Thg1)-Like Protein, with Nucleotide in the Activation and Nucleotidyl Transfer Sites. PLoS One. 2013 Jul 3;8(7):e67465. doi: 10.1371/journal.pone.0067465. Print 2013. PMID:23844012 doi:10.1371/journal.pone.0067465
- ↑ Jackman JE, Gott JM, Gray MW. Doing it in reverse: 3'-to-5' polymerization by the Thg1 superfamily. RNA. 2012 May;18(5):886-99. doi: 10.1261/rna.032300.112. Epub 2012 Mar 28. PMID:22456265 doi:http://dx.doi.org/10.1261/rna.032300.112
- ↑ Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases. Proc Natl Acad Sci U S A. 2010 Nov 8. PMID:21059936 doi:10.1073/pnas.1010436107
- ↑ Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases. Proc Natl Acad Sci U S A. 2010 Nov 8. PMID:21059936 doi:10.1073/pnas.1010436107
- ↑ Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases. Proc Natl Acad Sci U S A. 2010 Nov 8. PMID:21059936 doi:10.1073/pnas.1010436107
- ↑ Nakamura A, Nemoto T, Heinemann IU, Yamashita K, Sonoda T, Komoda K, Tanaka I, Soll D, Yao M. Structural basis of reverse nucleotide polymerization. Proc Natl Acad Sci U S A. 2013 Dec 24;110(52):20970-5. doi:, 10.1073/pnas.1321312111. Epub 2013 Dec 9. PMID:24324136 doi:http://dx.doi.org/10.1073/pnas.1321312111
- ↑ http://www.capture.caltech.edu/