We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Cody Couperus/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 39: | Line 39: | ||
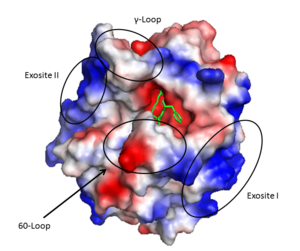
* Two surface patches referred to as exosite I and exosite II. | * Two surface patches referred to as exosite I and exosite II. | ||
| - | The <scene name='58/583418/ | + | The <scene name='58/583418/A_chain_nospin/1' target='0'>A chain</scene> is mostly helical and is wound around the B chain and shaped like a boomerang. It is bound to the B chain mostly through side chain interactions including a salt bridge and H-bond cluster at residues D14, E8, and E14c. Furthermore the C-terminus region forms a short amphipathic helix with hydrophobic side chains interacting with the B chain. |
The <scene name='58/583418/B_chain/1' target='0'>B chain</scene> contains the active site of the protein and has numerous notable structural features. The active site is formed at the rims of two interacting 6 stranded <scene name='58/583418/Beta_barrel/1'>beta barrel domains</scene> which are surrounded by 4 helical regions and many turns. | The <scene name='58/583418/B_chain/1' target='0'>B chain</scene> contains the active site of the protein and has numerous notable structural features. The active site is formed at the rims of two interacting 6 stranded <scene name='58/583418/Beta_barrel/1'>beta barrel domains</scene> which are surrounded by 4 helical regions and many turns. | ||
| Line 45: | Line 45: | ||
The serine protease <scene name='58/583418/Catalytic_triad/1' target='0'>catalytic triad</scene>, based on chymotrypsin numbering, are Ser195, His57, and Asp102. As is common with serine proteases, an oxanion hole is formed by backbone amides of Ser195 and Gly193. This has the functional role of stabilizing the oxanion intermediate involved in the serine protease mechanism. In addition, since thrombin cleaves after Arg/Lys the S1 specificity site, formed by the 180s- and 220s- loops, has Asp189 at the base to form a salt bridge with the incoming substrate. Furthermore, the S4 binding pocket accommodates hydrophobic substrate residues. | The serine protease <scene name='58/583418/Catalytic_triad/1' target='0'>catalytic triad</scene>, based on chymotrypsin numbering, are Ser195, His57, and Asp102. As is common with serine proteases, an oxanion hole is formed by backbone amides of Ser195 and Gly193. This has the functional role of stabilizing the oxanion intermediate involved in the serine protease mechanism. In addition, since thrombin cleaves after Arg/Lys the S1 specificity site, formed by the 180s- and 220s- loops, has Asp189 at the base to form a salt bridge with the incoming substrate. Furthermore, the S4 binding pocket accommodates hydrophobic substrate residues. | ||
| - | The '''active site''' cleft rims are formed by the hydrophobic and rigid 60-loop | + | The '''active site''' cleft rims are formed by the hydrophobic and rigid <scene name='58/583418/60_loop_ribbon/1'>60-loop</scene> (residues L60, Y60a, P60b, P60c, W60d, D60e, K60f, N60g, F60h, T60i, and N60g) and the γ-loop (residues T147, W147a, T147b, A147c, N147d, and V147f) while the base is mostly hydrophilic negatively charged amino acids. The cleft is deep compared to more promiscuous serine proteases, consequently substrates must either have a large loop that is cleaved or have favorable interactions with the insertion loops <ref>PMID: 16102053</ref>. |
Many other loops project out of the B chain but most are rigid due to proline and tryptophan residues. | Many other loops project out of the B chain but most are rigid due to proline and tryptophan residues. | ||
Revision as of 06:07, 28 April 2014
Thrombin: Structure and Function
| |||||||||||