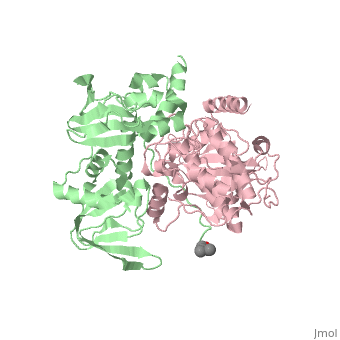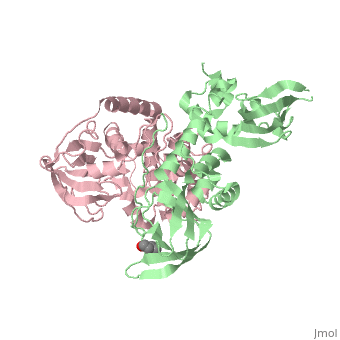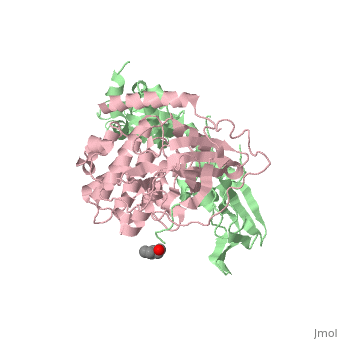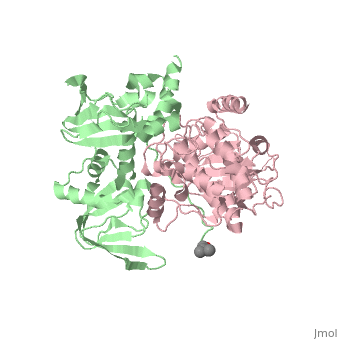Protein Kinase A
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
== Mechanism == | == Mechanism == | ||
PKA consists of a regulatory dimer, which each regulatory subunit attached to a catalytic subunit. When cAMP levels are low, the enzyme is intact, forming the R2C2 complex. However, when cAMP levels are high, two cAMP molecules bind to the regulatory sites of the complex, which leads to the dissociation of the R2C2 complex into an R2 subunit and two C subunits. When cAMP binds to the R chains, the binding allosterically removes the pseudostubstrate sequences, Arg-Arg-Gly-Ala-Ile, out of the catalytic sites. This psuedosubstrate sequence of R occupies the catalytic site of C and prevents it from interacting with protein substrates. The only difference between the pseudosubstrate sequence and the sequence for phophorylations is the alanine in place of the serine. The pseudosubstrate binds to PKA through specific interactions. The guanidinium group of the first arginine forms an ion pair with the carboxylate groups of the glutamate residue, Glu 127, of the enzyme, forming a salt bridge. In the same way, the second arginine interacts with two other carboxylate groups, forming two more salt bridges. Two leucine residues of the enzyme form a hydrophobic groove in which the nonpolar side chain of isoleucine fits into. The scene is shown here. Once released, the two enzymatically active C subunites are free to phosphorylate proteins. | PKA consists of a regulatory dimer, which each regulatory subunit attached to a catalytic subunit. When cAMP levels are low, the enzyme is intact, forming the R2C2 complex. However, when cAMP levels are high, two cAMP molecules bind to the regulatory sites of the complex, which leads to the dissociation of the R2C2 complex into an R2 subunit and two C subunits. When cAMP binds to the R chains, the binding allosterically removes the pseudostubstrate sequences, Arg-Arg-Gly-Ala-Ile, out of the catalytic sites. This psuedosubstrate sequence of R occupies the catalytic site of C and prevents it from interacting with protein substrates. The only difference between the pseudosubstrate sequence and the sequence for phophorylations is the alanine in place of the serine. The pseudosubstrate binds to PKA through specific interactions. The guanidinium group of the first arginine forms an ion pair with the carboxylate groups of the glutamate residue, Glu 127, of the enzyme, forming a salt bridge. In the same way, the second arginine interacts with two other carboxylate groups, forming two more salt bridges. Two leucine residues of the enzyme form a hydrophobic groove in which the nonpolar side chain of isoleucine fits into. The scene is shown here. Once released, the two enzymatically active C subunites are free to phosphorylate proteins. | ||
| + | PKA is inactivated by a feedback mechanism. PKA activates phosphodiesterase, which converts cAMP back to ATP. This reduces the amount of cAMP that can activate PKA. | ||
| - | == Disease == | ||
| - | |||
| - | == Relevance == | ||
| - | |||
| - | == Structural highlights == | ||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
| Line 24: | Line 18: | ||
[1]Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644 | [1]Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644 | ||
[2]C. Kim, N.-H. Xuong, and S.S. Taylor, "Crystal structure of a complex between catalytic and regulatory (Rlα) subunits of PKA,"Science 307, 690 (2005) | [2]C. Kim, N.-H. Xuong, and S.S. Taylor, "Crystal structure of a complex between catalytic and regulatory (Rlα) subunits of PKA,"Science 307, 690 (2005) | ||
| - | + | doi:10.2210/rcsb_pdb/mom_2012_8 | |
Revision as of 16:59, 29 April 2014
Protein kinase A (PKA)
| |||||||||||
References
[1]Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644 [2]C. Kim, N.-H. Xuong, and S.S. Taylor, "Crystal structure of a complex between catalytic and regulatory (Rlα) subunits of PKA,"Science 307, 690 (2005) doi:10.2210/rcsb_pdb/mom_2012_8