We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Cody Couperus/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 66: | Line 66: | ||
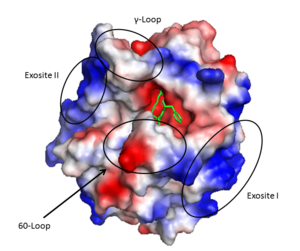
'''Exosite I''' is located on the B chain and had both basic and hydrophobic character. It is important in binding <scene name='58/583418/Fibrinogen/1'>fibrinogen</scene>, platelet activated receptors, and <scene name='58/583418/Thrombomodulin/1'>thrombomodulin</scene>. | '''Exosite I''' is located on the B chain and had both basic and hydrophobic character. It is important in binding <scene name='58/583418/Fibrinogen/1'>fibrinogen</scene>, platelet activated receptors, and <scene name='58/583418/Thrombomodulin/1'>thrombomodulin</scene>. | ||
| - | '''Exosite II''' is also part of the B chain, and derived from numerous basic amino acids, this is the site of <scene name='58/583418/Antithrombin/1'>heparin binding</scene> through the sulfate groups on the glycosaminoglycan. It is also the site of | + | '''Exosite II''' is also part of the B chain, and derived from numerous basic amino acids, this is the site of <scene name='58/583418/Antithrombin/1'>heparin binding</scene> through the sulfate groups on the glycosaminoglycan. It is also the site of GpIbα binding on the platelet surface. |
The <scene name='58/583418/Sodium_binding_loop/1'>sodium binding site</scene> is formed by the 180s- and 220s- loops. Na+ is bound by the backbone oxygens of Arg221a and Lys224 in addition to four water molecules in a classic [http://chemwiki.ucdavis.edu/Inorganic_Chemistry/Crystal_Field_Theory/High_Spin_and_Low_Spin_Complexes#Octahedral_Geometry octahedral geometry]<ref>PMID: 9108691</ref>. Through the covelent disulfide linkage between Cys220 and Cys 191 the sodium binding site is linked to Ser195 and the oxyanion hole. | The <scene name='58/583418/Sodium_binding_loop/1'>sodium binding site</scene> is formed by the 180s- and 220s- loops. Na+ is bound by the backbone oxygens of Arg221a and Lys224 in addition to four water molecules in a classic [http://chemwiki.ucdavis.edu/Inorganic_Chemistry/Crystal_Field_Theory/High_Spin_and_Low_Spin_Complexes#Octahedral_Geometry octahedral geometry]<ref>PMID: 9108691</ref>. Through the covelent disulfide linkage between Cys220 and Cys 191 the sodium binding site is linked to Ser195 and the oxyanion hole. | ||
Revision as of 15:17, 30 April 2014
| |||||||||||
References
- ↑ Fenton JW 2nd. Thrombin specificity. Ann N Y Acad Sci. 1981;370:468-95. PMID:7023326
- ↑ 2.0 2.1 Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000 Sep 14;407(6801):258-64. PMID:11001069 doi:http://dx.doi.org/10.1038/35025229
- ↑ Crawley JT, Lam JK, Rance JB, Mollica LR, O'Donnell JS, Lane DA. Proteolytic inactivation of ADAMTS13 by thrombin and plasmin. Blood. 2005 Feb 1;105(3):1085-93. Epub 2004 Sep 23. PMID:15388580 doi:http://dx.doi.org/10.1182/blood-2004-03-1101
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Lane DA, Philippou H, Huntington JA. Directing thrombin. Blood. 2005 Oct 15;106(8):2605-12. Epub 2005 Jun 30. PMID:15994286 doi:http://dx.doi.org/10.1182/blood-2005-04-1710
- ↑ Takagi T, Doolittle RF. Amino acid sequence studies on factor XIII and the peptide released during its activation by thrombin. Biochemistry. 1974 Feb 12;13(4):750-6. PMID:4811064
- ↑ Miljic P, Heylen E, Willemse J, Djordjevic V, Radojkovic D, Colovic M, Elezovic I, Hendriks D. Thrombin activatable fibrinolysis inhibitor (TAFI): a molecular link between coagulation and fibrinolysis. Srp Arh Celok Lek. 2010 Jan;138 Suppl 1:74-8. PMID:20229688
- ↑ 7.0 7.1 7.2 7.3 Huntington JA. Natural inhibitors of thrombin. Thromb Haemost. 2014 Apr 1;111(4):583-9. doi: 10.1160/TH13-10-0811. Epub 2014 Jan, 30. PMID:24477356 doi:http://dx.doi.org/10.1160/TH13-10-0811
- ↑ 8.0 8.1 8.2 8.3 8.4 Huntington JA. Thrombin inhibition by the serpins. J Thromb Haemost. 2013 Jun;11 Suppl 1:254-64. doi: 10.1111/jth.12252. PMID:23809129 doi:http://dx.doi.org/10.1111/jth.12252
- ↑ Esmon CT. The regulation of natural anticoagulant pathways. Science. 1987 Mar 13;235(4794):1348-52. PMID:3029867
- ↑ Kalafatis M, Rand MD, Mann KG. The mechanism of inactivation of human factor V and human factor Va by activated protein C. J Biol Chem. 1994 Dec 16;269(50):31869-80. PMID:7989361
- ↑ 11.0 11.1 Lu D, Kalafatis M, Mann KG, Long GL. Comparison of activated protein C/protein S-mediated inactivation of human factor VIII and factor V. Blood. 1996 Jun 1;87(11):4708-17. PMID:8639840
- ↑ Duga S, Asselta R, Tenchini ML. Coagulation factor V. Int J Biochem Cell Biol. 2004 Aug;36(8):1393-9. PMID:15147718 doi:http://dx.doi.org/10.1016/j.biocel.2003.08.002
- ↑ Saenko EL, Shima M, Sarafanov AG. Role of activation of the coagulation factor VIII in interaction with vWf, phospholipid, and functioning within the factor Xase complex. Trends Cardiovasc Med. 1999 Oct;9(7):185-92. PMID:10881749
- ↑ Camire, R. M. (2010). Platelet factor V to the rescue. Blood, 115(4), 753-754. DOI: 10.1182/blood-2009-11-252619
- ↑ Berkner KL. Vitamin K-dependent carboxylation. Vitam Horm. 2008;78:131-56. doi: 10.1016/S0083-6729(07)00007-6. PMID:18374193 doi:http://dx.doi.org/10.1016/S0083-6729(07)00007-6
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 16.6 16.7 Lechtenberg BC, Freund SM, Huntington JA. An ensemble view of thrombin allostery. Biol Chem. 2012 Sep;393(9):889-98. doi: 10.1515/hsz-2012-0178. PMID:22944689 doi:http://dx.doi.org/10.1515/hsz-2012-0178
- ↑ Tijburg PN, van Heerde WL, Leenhouts HM, Hessing M, Bouma BN, de Groot PG. Formation of meizothrombin as intermediate in factor Xa-catalyzed prothrombin activation on endothelial cells. The influence of thrombin on the reaction mechanism. J Biol Chem. 1991 Feb 25;266(6):4017-22. PMID:1995649
- ↑ Bobofchak KM, Pineda AO, Mathews FS, Di Cera E. Energetic and structural consequences of perturbing Gly-193 in the oxyanion hole of serine proteases. J Biol Chem. 2005 Jul 8;280(27):25644-50. Epub 2005 May 12. PMID:15890651 doi:http://dx.doi.org/10.1074/jbc.M503499200
- ↑ 19.0 19.1 19.2 19.3 Bode W, Mayr I, Baumann U, Huber R, Stone SR, Hofsteenge J. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J. 1989 Nov;8(11):3467-75. PMID:2583108
- ↑ Page MJ, Di Cera E. Evolution of peptidase diversity. J Biol Chem. 2008 Oct 31;283(44):30010-4. doi: 10.1074/jbc.M804650200. Epub 2008 , Sep 3. PMID:18768474 doi:http://dx.doi.org/10.1074/jbc.M804650200
- ↑ Schechter I, Berger A. On the size of the active site in proteases. I. Papain. 1967. Biochem Biophys Res Commun. 2012 Aug 31;425(3):497-502. doi:, 10.1016/j.bbrc.2012.08.015. PMID:22925665 doi:http://dx.doi.org/10.1016/j.bbrc.2012.08.015
- ↑ Huntington JA. Molecular recognition mechanisms of thrombin. J Thromb Haemost. 2005 Aug;3(8):1861-72. PMID:16102053 doi:http://dx.doi.org/10.1111/j.1538-7836.2005.01363.x
- ↑ Zhang E, Tulinsky A. The molecular environment of the Na+ binding site of thrombin. Biophys Chem. 1997 Jan 31;63(2-3):185-200. PMID:9108691
- ↑ Li W, Johnson DJ, Esmon CT, Huntington JA. Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat Struct Mol Biol. 2004 Sep;11(9):857-62. Epub 2004 Aug 15. PMID:15311269 doi:10.1038/nsmb811
- ↑ Spronk HM, Borissoff JI, ten Cate H. New insights into modulation of thrombin formation. Curr Atheroscler Rep. 2013 Nov;15(11):363. doi: 10.1007/s11883-013-0363-3. PMID:24026641 doi:http://dx.doi.org/10.1007/s11883-013-0363-3