Fatty acid amide hydrolase
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
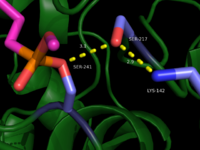
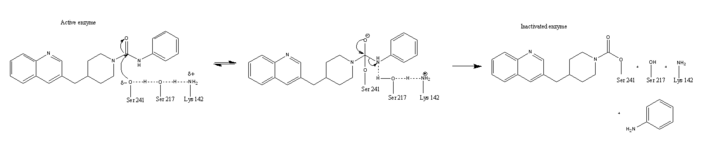
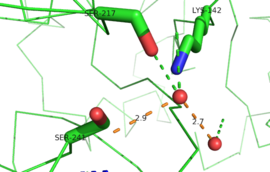
This evidence of convergent evolution between FAAH and other amidase signature enzymes supports the [http://euch6f.chem.emory.edu/burgidunitz.html Bürgi-Dunitz theory]. The Bürgi-Dunitz theory proposes that nucleophiles tend to follow a specific trajectory when attacking a carbonyl, resulting in many enzyme mechanisms having the same angle between an incoming nucleophile and the carbonyl it attacks [http://www.nature.com/nrd/journal/v11/n4/images/nrd3673-f4.jpg (FAAH bound to inhibitor)]. Water molecules in the active sites of enzymes are specifically positioned to force the nucleophile to approach at the exact [http://3.bp.blogspot.com/-NvsQyVPnLIw/UO91-BQTVgI/AAAAAAAAExw/-seGZcjU3DE/s400/burgi-duntz+trajectory.png "Bürgi-Dunitz angle"] of 107°. The determination that FAAH also has water molecules in its active site, helping the nucleophile to attack the amide carbonyl at a specific angle, adds additional support to the Bürgi-Dunitz theory <ref name="3LJ6"/>. | This evidence of convergent evolution between FAAH and other amidase signature enzymes supports the [http://euch6f.chem.emory.edu/burgidunitz.html Bürgi-Dunitz theory]. The Bürgi-Dunitz theory proposes that nucleophiles tend to follow a specific trajectory when attacking a carbonyl, resulting in many enzyme mechanisms having the same angle between an incoming nucleophile and the carbonyl it attacks [http://www.nature.com/nrd/journal/v11/n4/images/nrd3673-f4.jpg (FAAH bound to inhibitor)]. Water molecules in the active sites of enzymes are specifically positioned to force the nucleophile to approach at the exact [http://3.bp.blogspot.com/-NvsQyVPnLIw/UO91-BQTVgI/AAAAAAAAExw/-seGZcjU3DE/s400/burgi-duntz+trajectory.png "Bürgi-Dunitz angle"] of 107°. The determination that FAAH also has water molecules in its active site, helping the nucleophile to attack the amide carbonyl at a specific angle, adds additional support to the Bürgi-Dunitz theory <ref name="3LJ6"/>. | ||
| + | |||
==Applications== | ==Applications== | ||
Revision as of 18:58, 3 May 2014
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002 Nov 29;298(5599):1793-6. PMID:12459591 doi:10.1126/science.1076535
- ↑ 2.0 2.1 2.2 2.3 Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, Lazerwith S, Stiff C, Kamtekar S, Bhattacharya K, Zhang Y, Swaney S, Van Becelaere K, Stevens RC, Cravatt BF. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem Biol. 2009 Apr 24;16(4):411-20. PMID:19389627 doi:10.1016/j.chembiol.2009.02.013
- ↑ 3.0 3.1 3.2 3.3 Mileni M, Johnson DS, Wang Z, Everdeen DS, Liimatta M, Pabst B, Bhattacharya K, Nugent RA, Kamtekar S, Cravatt BF, Ahn K, Stevens RC. Structure-guided inhibitor design for human FAAH by interspecies active site conversion. Proc Natl Acad Sci U S A. 2008 Sep 2;105(35):12820-4. Epub 2008 Aug 27. PMID:18753625
- ↑ Mileni M, Garfunkle J, DeMartino JK, Cravatt BF, Boger DL, Stevens RC. Binding and inactivation mechanism of a humanized fatty acid amide hydrolase by alpha-ketoheterocycle inhibitors revealed from cocrystal structures. J Am Chem Soc. 2009 Aug 5;131(30):10497-506. PMID:19722626 doi:10.1021/ja902694n
- ↑ 5.0 5.1 5.2 Mileni M, Kamtekar S, Wood DC, Benson TE, Cravatt BF, Stevens RC. Crystal structure of fatty acid amide hydrolase bound to the carbamate inhibitor URB597: discovery of a deacylating water molecule and insight into enzyme inactivation. J Mol Biol. 2010 Jul 23;400(4):743-54. Epub 2010 May 21. PMID:20493882 doi:10.1016/j.jmb.2010.05.034
Similar Proteopedia Pages
Student Contributors
- Rachel Erkilla
- Melissa Jones
- Daniel Lange
- Carter Sharp
3D Structures of fatty acid amide hydrolase
Updated on 03-May-2014
3qj8 – rFAAH - rat
1mt5, 3qj9, 3qkv, 4hbp – rFAAH + inhibitor
2vya, 2wap, 2wj1, 2wj2, 3k7f, 3k83, 3k84, 3lj6, 3lj7, 3qk5, 3oj8, 3ppm, 3pr0, 4j5p – rFAAH (mutant) + inhibitor
4do3 – rFAAH + anti-inflammatory drug
Proteopedia Page Contributors and Editors (what is this?)
R. Jeremy Johnson, Michal Harel, Alexander Berchansky, Angel Herraez