This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2x0n
From Proteopedia
(Difference between revisions)
m (Protected "2x0n" [edit=sysop:move=sysop]) |
|||
| Line 1: | Line 1: | ||
| - | [[ | + | ==STRUCTURE OF GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM TRYPANOSOMA BRUCEI DETERMINED FROM LAUE DATA== |
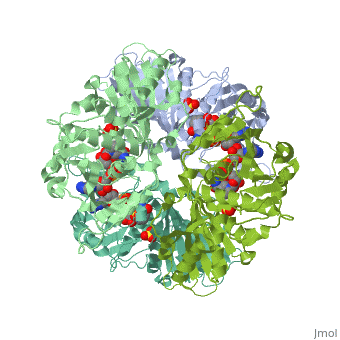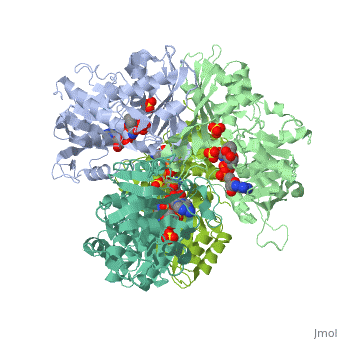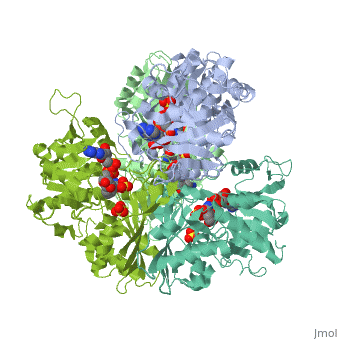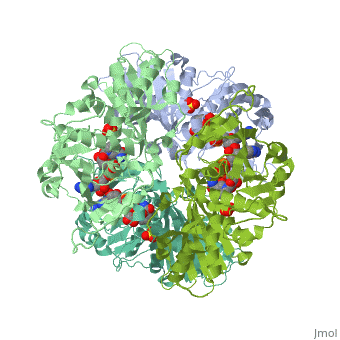
| + | <StructureSection load='2x0n' size='340' side='right' caption='[[2x0n]], [[Resolution|resolution]] 3.20Å' scene=''> | ||
| + | == Structural highlights == | ||
| + | <table><tr><td colspan='2'>[[2x0n]] is a 6 chain structure with sequence from [http://en.wikipedia.org/wiki/Trypanosoma_brucei_brucei Trypanosoma brucei brucei]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=1gga 1gga]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2X0N OCA]. <br> | ||
| + | </td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=NAD:NICOTINAMIDE-ADENINE-DINUCLEOTIDE'>NAD</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene><br> | ||
| + | <tr><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Glucokinase Glucokinase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.7.1.2 2.7.1.2] </span></td></tr> | ||
| + | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2x0n FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2x0n OCA], [http://www.rcsb.org/pdb/explore.do?structureId=2x0n RCSB], [http://www.ebi.ac.uk/pdbsum/2x0n PDBsum]</span></td></tr> | ||
| + | <table> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/x0/2x0n_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | The three-dimensional structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase [D-glyceraldehyde-3-phosphate:NAD+ oxidoreductase (phosphorylating), EC 1.12.1.12] from the sleeping-sickness parasite Trypanosoma brucei was solved by molecular replacement at 3.2-A resolution with an x-ray data set collected by the Laue method. For data collection, three crystals were exposed to the polychromatic synchrotron x-ray beam for a total of 20.5 sec. The structure was solved by using the Bacillus stearothermophilus enzyme model [Skarzynski, T., Moody, P. C. E. & Wonacott, A. J. (1987) J. Mol. Biol. 193, 171-187] with a partial data set which was 37% complete. The crystals contain six subunits per asymmetric unit, which allowed us to overcome the absence of > 60% of the reflections by 6-fold density averaging. After molecular dynamics refinement, the current molecular model has an R factor of 17.6%. Comparing the structure of the trypanosome enzyme with that of the homologous human muscle enzyme, which was determined at 2.4-A resolution, reveals important structural differences in the NAD binding region. These are of great interest for the design of specific inhibitors of the parasite enzyme. | ||
| - | + | Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data.,Vellieux FM, Hajdu J, Verlinde CL, Groendijk H, Read RJ, Greenhough TJ, Campbell JW, Kalk KH, Littlechild JA, Watson HC, et al. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2355-9. PMID:8460146<ref>PMID:8460146</ref> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | == References == | |
| - | + | <references/> | |
| - | + | __TOC__ | |
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
[[Category: Trypanosoma brucei brucei]] | [[Category: Trypanosoma brucei brucei]] | ||
[[Category: Hajdu, J.]] | [[Category: Hajdu, J.]] | ||
Revision as of 07:51, 14 May 2014
STRUCTURE OF GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM TRYPANOSOMA BRUCEI DETERMINED FROM LAUE DATA
| |||||||||||