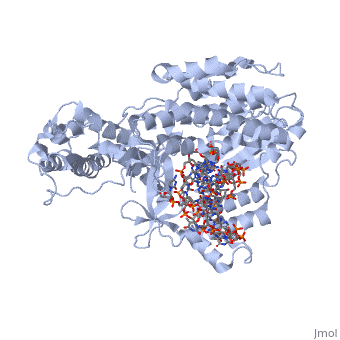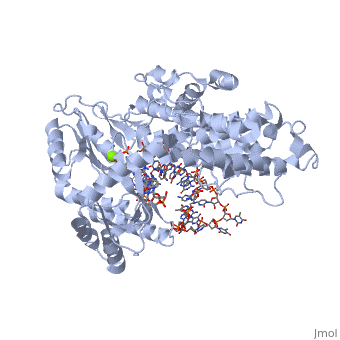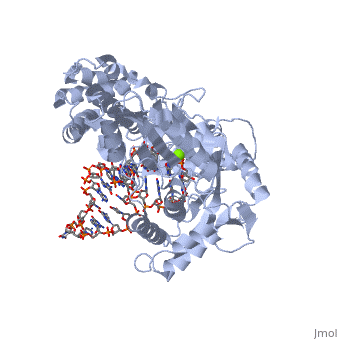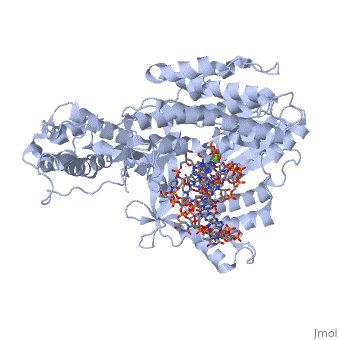
Catalytic Subunit of T. Castaneum TERT Polymerase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='3kyl' size='450' side='right' scene='' caption=''> | ||
=Introduction= | =Introduction= | ||
| Line 4: | Line 5: | ||
TERT is just one subunit of the Telomerase protein complex. TERT acts in junction with the TEN complex and a protein known as dyskerin in a homodimer. Each of the two parts of the dimer contains one TERT, one TEN, and one dyskerin molecule. Shown here is one TERT protein. | TERT is just one subunit of the Telomerase protein complex. TERT acts in junction with the TEN complex and a protein known as dyskerin in a homodimer. Each of the two parts of the dimer contains one TERT, one TEN, and one dyskerin molecule. Shown here is one TERT protein. | ||
| - | |||
| - | |||
| - | |||
| - | <StructureSection load='3kyl' size='500' frame='true' align='right' caption='Catalytic Subunit of TERT Polymerase bound to RNA Promoter, DNA Template Strand and Mg+2 ion, [[3kyl]]' [[3kyl]]> | ||
| - | |||
=Structural Overview= | =Structural Overview= | ||
| Line 46: | Line 42: | ||
Comparisons between the substrate bound TERT molecule and the substrate free molecule show small rigid body conformational changes in the positions of the subunits of the enzyme. These small conformational changes translate to a 3.5 angstrom decrease in the diameter of the interior cavity of the closed ring. The precise role of this conformational change is not known, although it may have to do with RNA binding ''in vivo'' | Comparisons between the substrate bound TERT molecule and the substrate free molecule show small rigid body conformational changes in the positions of the subunits of the enzyme. These small conformational changes translate to a 3.5 angstrom decrease in the diameter of the interior cavity of the closed ring. The precise role of this conformational change is not known, although it may have to do with RNA binding ''in vivo'' | ||
| - | </ | + | ===Structural Similarity to HIV Reverse Transcriptase=== |
| + | <scene name='52/522606/Cv/1'>HIV-1 reverse transcriptase (grey, green) complex with RNA, DNA, antibody heavy chain (yellow), light chain (pink)</scene> ([[1hys]]). | ||
| - | |||
| - | |||
| - | |||
| - | {{STRUCTURE_1hys| PDB=1hys | SIZE=300| SCENE= |left|CAPTION= HIV-1 reverse transcriptase (grey, green) complex with RNA, DNA, antibody heavy chain (yellow), light chain (pink) [[1hys]].}} | ||
| - | |||
| - | ===Structural Similarity to HIV Reverse Transcriptase=== | ||
It has been postulated that the mechanism by which TERT performs elongation of the telomeres is similar to the mechanism used by reverse transcriptases such as those found in retroviruses like HIV. Comparing the two proteins, it can be seen that there exist subtle differences in the structure, but the residues of the active sites are highly conserved across the two types of proteins, and the structural domains remain the same across the two as well. For example, the thumb loop of both is responsible for guiding the DNA substrate into the binding pocket of the enzyme, and orienting it in the correct direction. The Tyr256 and Gln308 of the nucleotide binding pocket are also found in HIV-RT. <ref name="Main"/> | It has been postulated that the mechanism by which TERT performs elongation of the telomeres is similar to the mechanism used by reverse transcriptases such as those found in retroviruses like HIV. Comparing the two proteins, it can be seen that there exist subtle differences in the structure, but the residues of the active sites are highly conserved across the two types of proteins, and the structural domains remain the same across the two as well. For example, the thumb loop of both is responsible for guiding the DNA substrate into the binding pocket of the enzyme, and orienting it in the correct direction. The Tyr256 and Gln308 of the nucleotide binding pocket are also found in HIV-RT. <ref name="Main"/> | ||
| - | [[Image:Difference_between_HIVRT_and_TERT.png | | + | [[Image:Difference_between_HIVRT_and_TERT.png|left|450px|thumb]] |
| - | + | ||
| - | + | ||
| - | + | ||
=Current Research= | =Current Research= | ||
| Line 75: | Line 63: | ||
TERT Polymerase has been shown to be inhibited by the human protein PinX1. PinX1 works by binding to the site where the RNA template is usually bound. The PinX1 protein is usually found in the nucleolus and nucleoplasm in humans, but in cancer cells TERT is found in the nucleoplasm. It is postulated that PinX1 acts as a tumor suppressor for this reason. <ref>FEBS Letters, Volume 579, Issue 4, 7 February 2005, Pages 859–862 http://dx.doi.org/10.1016/j.febslet.2004.11.036</ref> | TERT Polymerase has been shown to be inhibited by the human protein PinX1. PinX1 works by binding to the site where the RNA template is usually bound. The PinX1 protein is usually found in the nucleolus and nucleoplasm in humans, but in cancer cells TERT is found in the nucleoplasm. It is postulated that PinX1 acts as a tumor suppressor for this reason. <ref>FEBS Letters, Volume 579, Issue 4, 7 February 2005, Pages 859–862 http://dx.doi.org/10.1016/j.febslet.2004.11.036</ref> | ||
| - | + | </StructureSection> | |
===3D structures of TERT polymerase=== | ===3D structures of TERT polymerase=== | ||
Revision as of 09:20, 22 May 2014
| |||||||||||
3D structures of TERT polymerase
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010 Apr;17(4):513-8. Epub 2010 Mar 28. PMID:20357774 doi:10.1038/nsmb.1777
- ↑ Bernardes de Jesus B, Vera E, Schneeberger K, Tejera AM, Ayuso E, Bosch F, Blasco MA. Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol Med. 2012 Aug;4(8):691-704. doi: 10.1002/emmm.201200245. Epub 2012 May, 15. PMID:22585399 doi:10.1002/emmm.201200245
- ↑ European Journal of Cancer, Volume 33, Issue 5, April 1997, Pages 787–791 http://dx.doi.org/10.1016/S0959-8049(97)00062-2
- ↑ FEBS Letters, Volume 579, Issue 4, 7 February 2005, Pages 859–862 http://dx.doi.org/10.1016/j.febslet.2004.11.036