From Proteopedia
(Difference between revisions)
proteopedia linkproteopedia link
|
m |
| Line 26: |
Line 26: |
| | ===The Chromophore=== | | ===The Chromophore=== |
| | | | |
| - | The <scene name='Green_Fluorescent_Protein/Chromophore/1'>chromophore</scene> (<scene name='Green_Fluorescent_Protein/1ema_gfp_chromophorezoom/6'>top view</scene>) of GFP is located at the center of the β-barrel with a wild-type excitation peak of 395 nm, and a minor peak at 475 nm (about three times less intense<ref name="Tsien" />) <ref name="Yang" /><ref name="Cubitt" /><ref name="Ormo" /><ref name="Phillips" /> with extinction coefficients of approximately 30,000 and 7,000 M<sup>-1</sup> cm<sup>-1</sup>, respectively.<ref name="Yang" /><ref name="Phillips" /> Interestingly, the ''Aequorea victoria'' jellyfish utilizes the smaller of the two excitation peaks as pure aequorin emits a light of 470 nm.<ref name="Tsien">Tsien, Roger Y. 1998. The Green Fluorescent Protein. Annual Review in Biochemistry. 67:509-544.</ref> The relative amplitudes of these two excitation peaks can vary depending on environmental factors and previous illumination.<ref name="Ormo" /> For example, continued excitation leads to a diminution of the 395 nm excitation peak with a reciprocal amplification of the 475 nm peak.<ref name="Phillips" /> Regardless of absorption, the chromophore of GFP emits light of 508 nm.<ref name="Yang" /><ref name="Cubitt" /><ref name="Ormo" /><ref name="Phillips" /> | + | The <scene name='10/100139/Chromophore/2'>chromophore</scene> (<scene name='10/100139/Green_fluorescent_protein/1'>top view</scene>) of GFP is located at the center of the β-barrel with a wild-type excitation peak of 395 nm, and a minor peak at 475 nm (about three times less intense<ref name="Tsien" />) <ref name="Yang" /><ref name="Cubitt" /><ref name="Ormo" /><ref name="Phillips" /> with extinction coefficients of approximately 30,000 and 7,000 M<sup>-1</sup> cm<sup>-1</sup>, respectively.<ref name="Yang" /><ref name="Phillips" /> Interestingly, the ''Aequorea victoria'' jellyfish utilizes the smaller of the two excitation peaks as pure aequorin emits a light of 470 nm.<ref name="Tsien">Tsien, Roger Y. 1998. The Green Fluorescent Protein. Annual Review in Biochemistry. 67:509-544.</ref> The relative amplitudes of these two excitation peaks can vary depending on environmental factors and previous illumination.<ref name="Ormo" /> For example, continued excitation leads to a diminution of the 395 nm excitation peak with a reciprocal amplification of the 475 nm peak.<ref name="Phillips" /> Regardless of absorption, the chromophore of GFP emits light of 508 nm.<ref name="Yang" /><ref name="Cubitt" /><ref name="Ormo" /><ref name="Phillips" /> |
| | | | |
| | Three amino residues in the central α-helix constitute the fluorophore of GFP: Ser<sup>65</sup>Tyr<sup>66</sup>Gly<sup>67</sup> (see below). Tsien et al. discovered that this tri-peptide sequence is post-translationally modified by internal cyclization and oxidation<ref name="Haldar" /> to produce a <scene name='Green_Fluorescent_Protein/Chromophore_structure/1'>4-(p-hydroxybenzylidene)-imidazolidin-5-one</scene> structure.<ref name="Yang" /> Studies with E. coli proposed a sequential mechanism for the formation of the fluorophore that was initiated by a rapid cyclization between Ser<sup>65</sup> and Gly<sup>67</sup> to form an imidazolin-5-one intermediate.<ref name="Yang" /> This rapid cyclization is carried out via nucleophilic attack of the amino group from Gly<sup>67</sup> on the carbonyl group of Ser<sup>65</sup> to form a five-membered ring. The loss of water then forms the imidazolin-5-one intermediate.<ref name="Cubitt" /> Cyclization is succeeded by a much slower rate-limiting oxygenation of the Tyr<sup>66</sup> hydroxybenzyl side chain by atmospheric oxygen (No fluorescence was seen in anaerobically grown E. coli.), resulting in the 4-(p-hydroxybenzylidene)-imidazolidin-5-one stucture.<ref name="Yang" /><ref name="Cubitt" /><ref name="Phillips" /> The double bond that results from this series of reactions results in the linkage of the two π-systems of the rings, forming a larger conjugated system essential for fluorophore stability. <ref name="Bublitz"> Bublitz G, King BA, Boxer SG. 1998. Electronic structure of the chromophore in green fluorescent protein (GFP). Journal of the American Chemical Society. 120(36): 9370-9371. DOI 10.1021/ja98160e.</ref> | | Three amino residues in the central α-helix constitute the fluorophore of GFP: Ser<sup>65</sup>Tyr<sup>66</sup>Gly<sup>67</sup> (see below). Tsien et al. discovered that this tri-peptide sequence is post-translationally modified by internal cyclization and oxidation<ref name="Haldar" /> to produce a <scene name='Green_Fluorescent_Protein/Chromophore_structure/1'>4-(p-hydroxybenzylidene)-imidazolidin-5-one</scene> structure.<ref name="Yang" /> Studies with E. coli proposed a sequential mechanism for the formation of the fluorophore that was initiated by a rapid cyclization between Ser<sup>65</sup> and Gly<sup>67</sup> to form an imidazolin-5-one intermediate.<ref name="Yang" /> This rapid cyclization is carried out via nucleophilic attack of the amino group from Gly<sup>67</sup> on the carbonyl group of Ser<sup>65</sup> to form a five-membered ring. The loss of water then forms the imidazolin-5-one intermediate.<ref name="Cubitt" /> Cyclization is succeeded by a much slower rate-limiting oxygenation of the Tyr<sup>66</sup> hydroxybenzyl side chain by atmospheric oxygen (No fluorescence was seen in anaerobically grown E. coli.), resulting in the 4-(p-hydroxybenzylidene)-imidazolidin-5-one stucture.<ref name="Yang" /><ref name="Cubitt" /><ref name="Phillips" /> The double bond that results from this series of reactions results in the linkage of the two π-systems of the rings, forming a larger conjugated system essential for fluorophore stability. <ref name="Bublitz"> Bublitz G, King BA, Boxer SG. 1998. Electronic structure of the chromophore in green fluorescent protein (GFP). Journal of the American Chemical Society. 120(36): 9370-9371. DOI 10.1021/ja98160e.</ref> |
Revision as of 18:43, 8 July 2014
| Green fluorescent protein (GFP) is a bioluminescent polypeptide consisting of 238 residues isolated from the body of Aequorea victoria jellyfish.[1] GFP converts the blue chemiluminescent of aequorin in the jellyfish into green fluorescent light.[2] It remains unclear why these jellyfish use fluorescence, why green is better than blue, or why they produce a separate protein for green fluorescence as opposed to simply mutating the present aequorin to shift its wavelength,[3] but in the laboratory, GFP can be incorporated into a variety of biological systems in order to function as a marker protein. Since its discovery in 1962, GFP has become a significant contributor to the research of monitoring gene expression, localization, mobility, traffic, interactions between various membrane and cytoplasmic proteins, as well as many others.See also Green Fluorescent Protein: Research Tool.[4]
History
Aequorea victoria was first discovered and investigated for its bioluminescence by Frank Johnson, who invited Osamu Shimomura to work with him in on a small island not far from British Columbia, where the jellyfish is abundant.[5] Found off the west coast of the United States between British Columbia and central California,[6] the jellyfish was considered a local phenomenon as it would drift in and out of the harbors.[5]
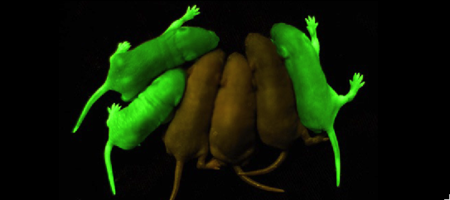 Mice with GFP inserted into their genomes for neurology studies. Shimomura was originally looking only to isolate the blue luminescent protein of Aequorea victoria, traditionally thought to be luciferase, but it would soon become apparent that the glow was in fact due to aequorin, a substance related, but slightly varying from luciferase.[4][5] However, the light emitted from aequorin still differed from the light emitted from the wild jellyfish. This quandary led to the discovery of the green fluorescent protein responsible for this disparity, but sufficient amounts of the protein could not be collected for study until 1979. The journey to discover the nature of GFP had begun.[5]
In the 1990’s, Douglas Prasher, Frank Predergast, and co-workers successfully cloned the gene that encoded for GFP. Martin Chalfie further pursued this line of work and was eventually able to express GFP in heterologous systems such as E. coli and C. elegans. Chalfie’s research provided the first evidence that GFP was unique as it did not require the presence of any exogenous substance or cofactor for fluorescence.[4] The lack for the need for a cofactor proved that the cloned GFP gene contained all the information necessary for posttranslational synthesis of the chromophore. [3]
Roger Tsien and co-workers were intrigued by the absence of a necessary cofactor and began to research the structure of GFP and how it relates to its fluorescence. They discovered that a helix within the beta barrel structure of GFP actually contained a fluorophore responsible for fluorescence. In researching its structure, they were able to develop GFP derivatives with improved fluorescence and photo-stability. Shimomura, Chalfie, and Tsien were each recognized for their work involving GFP with the Nobel Prize in 2008.[4] In the time since the work of these three researchers, GFP has been successfully expressed and utilized in bacteria, yeast, slime mold, plants, drosophila fruit flies, zebra-fish, and mammalian cells.[2] Below, mice have had GFP inserted into their genomes for studies in neurology.
Structure
Primary & Secondary Structure
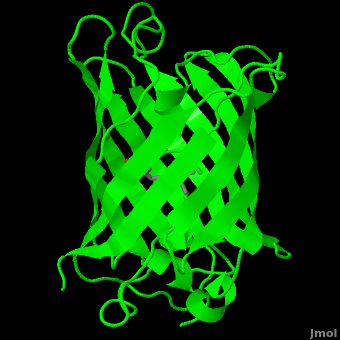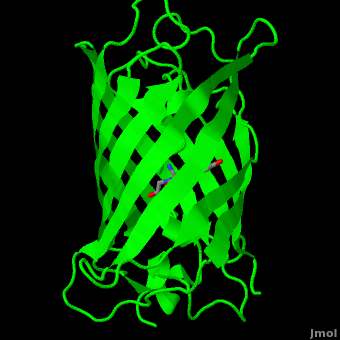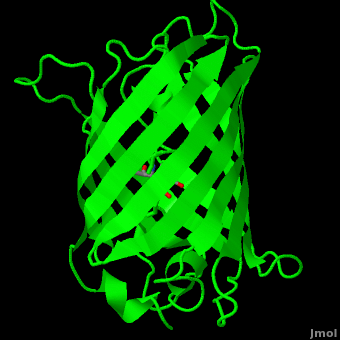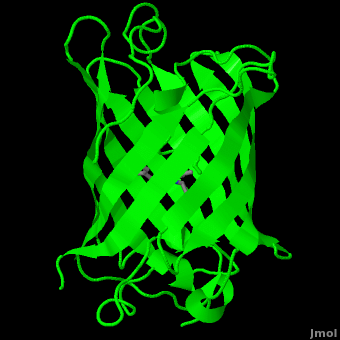
Green fluorescent protein () is a 21 kDa protein consisting of 238 residues strung together[7] to form a
of five α-helices and one eleven-stranded β-pleated sheet,[1] where each strand contains nine to thirteen residues each.[8] (To view the primary and secondary structure of GFP, go to .) These β-strands display an almost “seamless symmetry” in which only two of the strands vary in structural content.[9] This β-sheet conforms itself through regular hydrogen bonding into a β-barrel.[2] In GFP, the structure is so regular that of water molecules (red) can be seen following the structure of the barrel.[9] Together with the α-helices at either end of the molecule, a nearly perfect cylinder is produced, 42Å long and 24Å in diameter,[8] creating what is referred to as a “β-can” formation.[9] The short helical segments at either end of the cylinder form “caps” to further protect the interior of the β-barrel.[9] Overall stability is maintained by this β-can structure, helping to resist unfolding from heat and other denaturants.[2]
One can be found running through the central axis of the β-barrel,[4] roughly to the symmetry axis of the barrel.[8] This helix is extremely important as it contains the fluorophore responsible for fluorescence.[2][4] This α-helix in particular is highly stabilized by the many that are made with each strand of the barrel.[10]
.
The Chromophore
The () of GFP is located at the center of the β-barrel with a wild-type excitation peak of 395 nm, and a minor peak at 475 nm (about three times less intense[3]) [2][11][8][9] with extinction coefficients of approximately 30,000 and 7,000 M-1 cm-1, respectively.[2][9] Interestingly, the Aequorea victoria jellyfish utilizes the smaller of the two excitation peaks as pure aequorin emits a light of 470 nm.[3] The relative amplitudes of these two excitation peaks can vary depending on environmental factors and previous illumination.[8] For example, continued excitation leads to a diminution of the 395 nm excitation peak with a reciprocal amplification of the 475 nm peak.[9] Regardless of absorption, the chromophore of GFP emits light of 508 nm.[2][11][8][9]
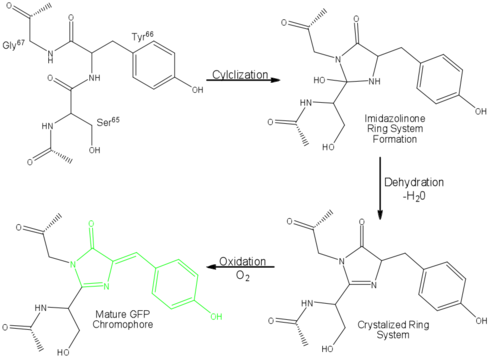
Three amino residues in the central α-helix constitute the fluorophore of GFP: Ser65Tyr66Gly67 (see below). Tsien et al. discovered that this tri-peptide sequence is post-translationally modified by internal cyclization and oxidation[4] to produce a structure.[2] Studies with E. coli proposed a sequential mechanism for the formation of the fluorophore that was initiated by a rapid cyclization between Ser65 and Gly67 to form an imidazolin-5-one intermediate.[2] This rapid cyclization is carried out via nucleophilic attack of the amino group from Gly67 on the carbonyl group of Ser65 to form a five-membered ring. The loss of water then forms the imidazolin-5-one intermediate.[11] Cyclization is succeeded by a much slower rate-limiting oxygenation of the Tyr66 hydroxybenzyl side chain by atmospheric oxygen (No fluorescence was seen in anaerobically grown E. coli.), resulting in the 4-(p-hydroxybenzylidene)-imidazolidin-5-one stucture.[2][11][9] The double bond that results from this series of reactions results in the linkage of the two π-systems of the rings, forming a larger conjugated system essential for fluorophore stability. [12]
The process is completely auto-catalytic such that there are no known co-factors or enzymatic components required.[2] Despite the stability of the final product, while the chromophore is forming, the environmental temperature cannot drop below 30°C or the yield of viable GFP will decrease substantially.[2][9] This, of course, is not an issue for the protein in nature as the jellyfish is unlikely to encounter waters of this degree in the Pacific Northwest.[3] Such a temperature sensitivity is only relevant during formation as the stability of the final product is maintained through a network of close contacts surrounding the fluorophore.[2] This, however, can and has been used in pulse-chase experiments in which the GFP-expressing cells are exposed to varying temperatures in place of labeled vs. unlabeled trials.[3]
As the central α-helix is not located directly in the center of the β-barrel, cavities of differing area exist on either side of the chromophore. The larger cavity, consisting of about 135 Å,[8] does not open out to the bulk solvent, but rather houses .[8][13] Had this space not been occupied, it would be expected to considerably destabilize the protein as a whole. The hydrogen bonding created by the presence of the water molecules, however, helps to link the buried of Glu222 and Gln69 that would otherwise be actively polar.[8] Therefore, the water molecules are extremely important in establishing a hydrogen bonding network about the chromophor.[14]
The opposite side of the chromophore, however, is within close proximity of several aromatic and polar side chains. Several between the surrounding residues and the chromophore are present including: hydrogen bonds of His148, Thr203, and Ser205 with the phenolic hydroxyl of Tyr66; Arg96 and Gln94 with the carbonyl of the imidazolidinone ring; and hydrogen bonds of Glu222 with the side chain of Thr65. Additional hydrogen bonding in the area around the chromophore helps to stabilize Arg96 in the protonated form, which suggests the presence of a partial negative charge on the carbonyl oxygen of the imidazolidinone ring in the deprotonated fluorophore.[8] Arg96 and Gln94 in turn help to steady the imidazolidone.[2] Therefore, it is thought that Arg96 is essential for the formation of the fluorophore by catalyzing the initial ring closure.[8] Tyr145 provides a stabilizing
[15] with the benzyl ring of the chromophore.[8] The stability provided by the internal polar interactions are further augmented by the surrounding β-barrel.
The β-barrel provides a highly constrained environment that protects the chromophore from the bulk solvent,[4] nearly creating the atmosphere of a vacuum.[14] This is most likely responsible for the small Stoke’s shift, or the small wavelength difference between excitation and emission.[8]
.
Findings show that fluorescence will not occur from a naked chromophore, but rather requires the protection of the β-can structure.[11] However, in crystallum GFP will exhibit a nearly identical fluorescence spectrum and lifetime when compared with aqueous GFP. These two elements point to a fluorescence that is not inherent to the isolated fluorophore,[2][9] but rather from the auto-catalytic cyclization of the polypeptide sequence Ser65Tyr66Gly67 and subsequent oxidation of Tyr66.[9] However, this sequence is found in many proteins - why does GFP fluoresce? According to Phillips (1997), fluorophore formation is due to the close proximity of the backbone atoms between Ser65. and Gly67 gained through a lack of sterical hindrance by the hydrogen atom side chain of glycine. In fact, no functional fluorescent proteins have been found in which any other amino acid other than glycine was found at position 67. Even so, there are still proteins that have this specific sequence, therefore, there must be another inherent property to GFP that is still left misunderstood.[9]
This quandary led Phillips to study the acid/base chemistry catalyzing the initial cyclization of the chromophore. He found that Arg96 actually acts as a
by withdrawing electrons through hydrogen bonding with the carbonyl oxygen of Ser65 to activate the carbonyl carbon for nucleophilic attack by the amide nitrogen of Gly67. This mechanism was further supported by ab initio calculations, as well as database searches of similar compounds and protein sequences. Through acid/base chemistry, the chromophore is stabilized by resonance.[9] Femtosecond Raman spectroscopy has been used to map the alteration of the structure close the chromophore during excited-state protein transfer and shown that chromophore wagging is orchestrated by the protein environment.[16]
Mutant Studies
Many mutant green fluorescent proteins have been developed in order to further understand the structure and mechanism of the fluorophore. The first mutagenesis studies simply
of the amino acid sequence (. NOTE: The structure represented here is already truncated at the carbonyl terminus). Shortening the polypeptide by more than seven amino acids from either terminus lead to a total loss of fluorescence, as well as a complete failure to absorb light at the traditional wavelengths. This is most likely due to the structure of the protein. The last seven amino acid residues of the carboxyl terminus are roughly disordered, and thus do not interfere with the overall structure. After seven residues, however, the capping α-helix structure is disrupted, leading to an unstable or unformed chromophore. The is less understood, but the same principle still applies even though the β-barrel does not begin until residue ten or eleven.[2]
Point mutations have also been extensively studied in order to examine their effects on the chromophore. In general, most point mutations lead to a diminished excitation, especially in regions of the sequence adjacent to the fluorophore or those that interact with the fluorophore. An exception to this trend is the Ser65Thr66 mutant (normal Ser65Tyr66), which actually increases fluorescence intensity, although the reason is unclear.[2]
An interesting mutation discovered by Ormo et al. (1996) was the Thr65Tyr66Gly67 mutant, which produces an α-helical conformation in the chromophore opposed to the normal conformation, which is nearly perpendicular to the helical axis, due to its interaction with Arg96. This further supports the idea that Arg96 is an important factor in the structural arrangement required for cyclization, perhaps by promoting the attack of Gly67 on the carbonyl carbon of Thr65.[8]
In high protein concentrations, GFP has been found to dimerize under the influence of high ionic strength between the two monomers. In Aequorea victoria, the aequorin is able to bind to the (1gfl), but not the monomer. Therefore, dimerization is a very important structural feature in terms of its function, as it also assists the GFP to absorb energy at the excitation wavelength of aequorin even though GFP has only a “modest” extinction coefficient. As a result, dimers, and often even higher (1w7s), are predominant protein populations within the jellyfish.[11]
.
Using GFP as a Research Tool
A description of some of the ways GFP is being used as a tool in research is at Green_Fluorescent_Protein:_Research_Tool.
|
3D Structures of Green Fluorescent Protein
Updated on 08-July-2014
2qu1, 2h9w – jGFP - jellyfish
3la1, 3i19, 2wur, 3gex, 2qrf, 2qt2, 2qz0, 2gj1, 2gj2, 3cb9, 3cbe, 3cd1, 3cd9, 2hjo, 2hqz, 2hrs, 2okw, 2oky, 2q57, 2due, 2duf, 2dug, 2duh, 2dui, 2q6p, 2hcg, 2hfc, 2hgd, 2hgy, 2awj, 2awk, 2awl, 2awm, 2g16, 2g2s, 2g3d, 2g5z, 2g6e, 2ah8, 2aha, 2fwq, 2fzu, 2b3p, 2b3q, 1z1p, 1z1q, 1yhg, 1yhh, 1yhi, 1yj2, 1yjf, 1s6z, 1q4a, 1q4b, 1q4c, 1q4d, 1q4e, 1q73, 1qyf, 1qyo, 1qyq, 1qst, 1qy3, 1cv7, 1jc0, 1jc1, 1jby, 1jbz, 1kp5, 1kyp, 1hcj, 1h6r, 1b9c, 1c4f, 1emc, 1eme, 1emf, 1emk, 1eml, 1emm, 2emd, 2emn, 2emo, 1emb, 1gfl, 1ema, 2y0g, 3gj1, 3gj2, 3p28, 1qxt, 3sry, 3ssp, 3st0, 3ufz, 3ug0, 4eul, 4ges, 4j88, 4j89, 4j8a, 3w1c, 3w1d, 4h47, 4h48, 4jfg, 4lqt, 4lqu, 4lw5 – jGFP (mutant)
3evp – jGFP circular permutation
2h6v – jGFP+imidazole derivative
1rm9, 1rmm, 1rmo, 1rmp, 1rrz – jGFP containing fluorotryptophan
2o24, 2o29, 2o2b, 1w7u, 1w7t, 1w7s, 1emg – jGFP (mutant)+imidazole derivative
1kyr – jGFP (mutant)+imidazole derivative+Cu
4gf6 - jGFP (mutant) +Cu
1kys – jGFP (mutant)+imidazole derivative+Zn
3ss0, 3ssh, 3ssk, 3ssl, 3sst, 3ssv, 3ssy, 3sv5, 3svb, 3svc, 3svd, 3sve – jGFP (mutant)+imidazole derivative+ halide
3ogo – jGFP+cGFP nanobody – camel
3g9a, 3k1k – jGFP+minimize nanobody – Lama pacos
2qle – GFP (mutant) – Azotobacter vinelandii
2rh7 – GFP – Renilla reniformis
3adf – monomeric azami green – Galaxea fascicularis
2vzx – GFP DENDRA2 – Dendronephthya
2gw3 – GFP KAEDE – Trachiphyllia geoffroyi
2pox, 2gx0, 2gx2, 2iov, 2ie2 – FP DRONPA – Echinophyllia
2dd7 – CpGFP - Chiridius poppei
2dd9 – CpGFP (mutant)
2c9i – saGFP – sea anemone
1xmz – saGFP (mutant)
2c9j – GFP – Cerianthus membranaceus
2hpw – GFP – Clytia gregaria
2g3o – PpGFP – Pontellina plumata
2g6x, 2g6y – PpGFP (mutant)
3lva, 3lvc, 3lvd – GFP (mutant) – Aequoarea coerulescens
4dkm, 4dkn – GFP – Florida lancelet
4hvf – GFP N terminal (mutant) – common lancelet
4dxi, 4dxm – GFP – synthetic
Yellow fluorescent protein
3dpw, 3dpx, 3dpz, 3dq1, 3dq2, 3dq3, 3dq4, 3dq5, 3dq6, 3dq7, 3dq8, 3dq9, 3dqa, 3dqc, 3dqd, 3dqe, 3dqf, 3dqh, 3dqi, 3dqj, 3dqk, 3dql, 3dqm, 3dqn, 3dqo, 3dqu, 1myw, 1huy, 2yfp, 1yfp, 3ed8, 3v3d – jGFP (mutant)
1f09, 1f0b – jGFP (mutant)+imidazole derivative+I
2ogr – Z-FP - Zoanthus
2pxs, 2pxw, 1xa9, 1xae – Z-FP (mutant)
2jad – jGFP/glutaredoxin
Red fluorescent protein
2icr, 2ojk – Z-RFP
2fl1 – Z-RFP (mutant)
3bx9, 3bxa, 3bxb, 3bxc, 3e5t, 3e5w, 1uis, 3ip2, 3pj5, 3pj7, 3pjb, 3pib - EnRFP – Entacmaea quadricolor
3e5v, 3rwt – EnRFP (mutant)
1zgo, 2vad, 2vae, 1ggx – DiRFP – Discosoma
1zgp, 1zgq, 2h8q, 2v4e, 1g7k – DiRFP (mutant)
3cfa – AsRFP – Anemonia sulcata
3nt3, 3nt9 – RFP – artificial gene
1yzw – RFP – Heteractis
3ir8 – RFP – Montipora
4edo, 4eds – GFP – Entacmaea quadricolor
3u8a, 3u8c, 4kge, 4kgf, 4gob – GFP - synthetic
Cyan fluorescent protein
2wsn, 2wso, 2ydz - jGFP
2ye0, 2ye1, 3ztf, 4ar7, 4as8, 4b5y - jGFP (mutant)
2otb – cyan C-FP – Clavularia
2ote - cyan C-FP (mutant)
2zo6, 2zo7 – cyan FP – Fungia concinna
1oxd, 1oxe, 1oxf – cyan FP (mutant) – marker plasmid
Blue fluorescent protein
1bfp – jGFP (mutant)
4en1 – jGFP
Photoconvertible fluorescent protein
2vvh, 2vvi, 2vvj, 3p8u – LhGFP (mutant) – Lobophyllia hemprichii
1zux – LhGFP
2btj - LhGFP+imidazole derivative
2ddc, 1xss – FfFP – Favia favus
2ddd - FfFP (mutant)
3cff, 3cfh – AsGFP (mutant)
Green fluorescent protein chimera
3ai4 – jGFP/mPolymerase iota ubiquitin binding motif - mouse
3ai5, 3vht - jGFP/m ubiquitin
3o77, 3o78, 3ek4, 3ek7 - jGFP/myosin light chain kinase/calmodulin
4anj - jGFP/myosin light chain kinase + calmodulin
3evr, 3evu, 3evv - jGFP/myosin light chain kinase/calmodulin+Ca
3ek8, 3ekh, 3ekj, 3sg2, 3sg3, 3sg4, 3sg5, 3sg6, 3sg7, 3wlc, 3wld - jGFP/myosin light chain kinase/calmodulin (mutant)
3osq, 3osr – jGFP/maltose-binding protein
3u8p – jGFP/cytochrome b562
4jrb – jGFP/allergen BLA
4bdu – jGFP/apoptosis regulator BAX
4ik1, 4ik3, 4ik4, 4ik5, 4ik8, 4ik9 – jGFP/Rcamp
Reference for this Structure
Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.
References
- ↑ 1.0 1.1 [1], Protein Database (PDBsum): 1ema. European Bioinformatics (EBI); 2009.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 [2], Yang F, Moss LG, Phillips GN Jr. 1996. The molecular structure of green fluorescent protein. Biotechnology. 14: 1246-1251. DOI 10.1038/nbt1096-1246.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Tsien, Roger Y. 1998. The Green Fluorescent Protein. Annual Review in Biochemistry. 67:509-544.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 [3], Haldar S, Chattopadhyay A. 2009. The green journey. J Fluoresc. 19:1-2. DOI 10.1007/s10895-008-0455-6; biographical background on Douglas Prasher, Martin Chalfie and Roger Tsien.
- ↑ 5.0 5.1 5.2 5.3 [4], Shimomura O. The discovery of green fluorescent protein. Nobel Prize Lecture; 2009;; biographical background at Wikipedia.
- ↑ [5],Cowles D, Cowles J. Aequorea victoria. 2007. Walla Wall University.
- ↑ Primary structure at www.ebi.aci.uk.
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 9.12 9.13 Phillips GN Jr. Structure and dynamics of green fluorescent protein. Curr Opin Struct Biol. 1997 Dec;7(6):821-7. PMID:9434902
- ↑ Andrews BT, Gosavi S, Finke JM, Onuchic JN, Jennings PA. The dual-basin landscape in GFP folding. Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12283-8. Epub 2008 Aug 19. PMID:18713871
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 [6],Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien R. 1995. Understanding, improving, and using green fluorescent protein. Trends in Biochemical Sciences. 20(11): 448-455. DOI 0.1016/S0968-0004(00)89099-4.
- ↑ Bublitz G, King BA, Boxer SG. 1998. Electronic structure of the chromophore in green fluorescent protein (GFP). Journal of the American Chemical Society. 120(36): 9370-9371. DOI 10.1021/ja98160e.
- ↑ van Thor JJ, Sage, JT. 2006. Charge transfer in green fluorescent protein. Photochemical & Photobiological Sciences. 5:597-602. DOI 10.1039/b516525c.
- ↑ 14.0 14.1 Lammich L, Petersen MA, Nielsen MB, Andersen LH. The gas-phase absorption spectrum of a neutral GFP model chromophore. Biophys J. 2007 Jan 1;92(1):201-7. Epub 2006 Oct 13. PMID:17040991 doi:10.1529/biophysj.106.093674
- ↑ Information about edge-face (CH/π) interactions.
- ↑ Fang C, Frontiera RR, Tran R, Mathies RA. Mapping GFP structure evolution during proton transfer with femtosecond Raman spectroscopy. Nature. 2009 Nov 12;462(7270):200-4. PMID:19907490 doi:10.1038/nature08527
Additional Resources
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Wayne Decatur, Karsten Theis, Eran Hodis, Laura Carbone, Karl Oberholser, Mark Hoelzer, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky, Marius Mihasan, Joseph M. Steinberger, David Canner