User:Eric Martz/JSmol Notes
From Proteopedia
(→Speed) |
(→Speed) |
||
| Line 90: | Line 90: | ||
</td><td> | </td><td> | ||
900 | 900 | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | [[2bbn]] | ||
| + | </td><td> | ||
| + | 57 (2.7K x 21 NMR models) | ||
| + | </td><td> | ||
| + | 416 (20K x 20 NMR models) | ||
</td> | </td> | ||
</tr> | </tr> | ||
| Line 98: | Line 107: | ||
</table> | </table> | ||
| - | JSmol is significantly '''slower''' than Jmol. Rotation will be jerkier. Smaller molecules (< | + | JSmol is significantly '''slower''' than Jmol. Rotation will be jerkier. Smaller molecules (<10,000 atoms) are handled quite well. '''Large molecules''' (>20,000 atoms; see table at right) may be slow to load and process. (Sometimes I’ve seen delays of up to a minute, even more, during which JSmol freezes. These delays tend to happen with large molecules but are not simply related to size.) Multiple-model ensembles (such as from [[NMR]] experiments) may cause JSmol to freeze while taking minutes to load the ensemble, or loading may never finish. |
Here is how two websites that now use JSmol by default have handled these issues: | Here is how two websites that now use JSmol by default have handled these issues: | ||
Revision as of 22:52, 29 July 2014
Contents |
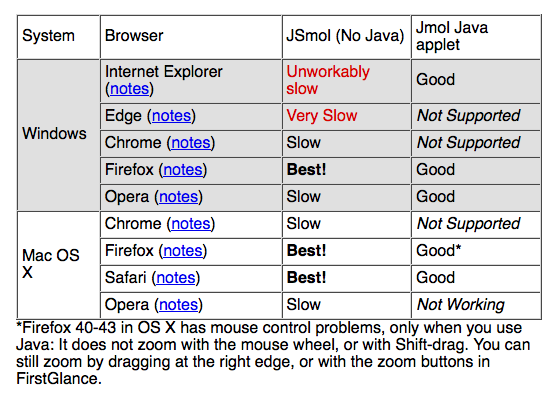
Browser Performance with JSmol
Different web browsers operate JSmol with very different performances. The findings below are based primarily on tests with FirstGlance in Jmol, and possibly results may be different for other JSmol websites.
Overview
The above snapshot is taken from the help in FirstGlance in Jmol: display any molecule in FirstGlance, then click on Trouble? (bottom of upper left panel) or Can't see the molecule? (bottom of right panel). The help appears in the lower left panel, and you will have to scroll down to see the above overview table.
Details
For more details, see the section Browser Pecularities in the Notes for FirstGlance.
Converting Websites from Jmol to JSmol
Things To Be Aware Of Before Converting To JSmol
Advantages
Of course there are huge advantages to using JSmol:
- Users of a JSmol website do not need to install Java (which is a likely security risk).
- A JSmol website will likely work on iPads and smart phones. (Java is not available for these platforms, so Jmol is not an option for them.)
Disadvantages
Speed
|
| ||
|
Atoms (Kilo) |
KD (Asymmetric Unit) |
|
|
1.7 |
28 |
|
|
6 |
87 |
|
|
12 |
160 |
|
|
23 |
233 |
|
|
45 |
826 |
|
|
60 |
900 |
|
|
57 (2.7K x 21 NMR models) |
416 (20K x 20 NMR models) |
|
|
Models with >10,000 atoms are in the largest ~10% of the PDB; with >20,000 atoms, the largest ~5%; with >50,000 atoms, the largest ~1.5% (counting all models for NMR ensembles). |
||
JSmol is significantly slower than Jmol. Rotation will be jerkier. Smaller molecules (<10,000 atoms) are handled quite well. Large molecules (>20,000 atoms; see table at right) may be slow to load and process. (Sometimes I’ve seen delays of up to a minute, even more, during which JSmol freezes. These delays tend to happen with large molecules but are not simply related to size.) Multiple-model ensembles (such as from NMR experiments) may cause JSmol to freeze while taking minutes to load the ensemble, or loading may never finish.
Here is how two websites that now use JSmol by default have handled these issues:
- FirstGlance in Jmol loads only the first model when there are more than one. It offers signed Jmol_S (Java) as an option (with a preference setting). The unsigned Jmol applet is no longer offered because it is deprecated and Oracle says soon it won't be allowed. FirstGlance recommends using Java when the molecule has >20,000 atoms, and even more strongly recommends it when >50,000 atoms.
- Proteopedia loads only amino acid alpha carbon atoms (or nucleic acid phosphorus atoms) and ligand atoms, since its ribbon view (secondary structure schematic) works fine without the other atoms. A prominent message is displayed that a simplified model is being shown, and there is a button to load the complete model. For registered users who are logged in, Proteopedia has a preference setting that will use Jmol_S instead of JSmol.
Browser Peculiarities
See the table above for an overview.
JSmol runs in the javascript of the browser (HTML5). Internet Explorer has unacceptably slow javascript (all versions, including version 11). It is the most widely used browser
1. Download Jmol. The download package includes both JSmol and the Jmol Java applet. Go to Jmol.Org, click Downloads.
2. Unzip the downloaded Jmol file. After unzipping, inside the main folder is jsmol.zip. Unzip that too.
3. Copy the jsmol folder (the one you unzipped in step 2) into the resource that you are upgrading. (You don’t need to copy any of the other files in the main folder.)
The following steps apply to the main html file of your resource:
4. Javascript files in the <header> section of the main html file. If the old page used Jmol.js, there will be a line like this that can be removed or commented out:
<!— COMMENT OUT THIS LINE <script src="jmol/Jmol.js"></script>
-->
Add these two lines. Jmol2.js replaces Jmol.js, defining the same functions you were already using in Jmol.js.
<script type="text/javascript" src="jsmol/JSmol.min.js"></script> <script type="text/javascript" src="jsmol/js/Jmol2.js"></script>
Other Conversion Advice
- Converting pages from Jmol to JSmol in the Jmol Wiki. This writeup is too technical for me to understand.