This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Journal:JBIC:26
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
[[Image:Scheme_1.png|left|450px|thumb|]] | [[Image:Scheme_1.png|left|450px|thumb|]] | ||
{{Clear}} | {{Clear}} | ||
| - | A strategy to control the activity of urease for medical and agricultural applications is to use enzyme inhibitors. Fluoride is a known urease inhibitor, but the structural basis of its mode of inhibition are still undetermined. Here, kinetic studies on the fluoride-induced inhibition of urease from ''Sporosarcina pasteurii'', a widespread and highly ureolytic soil bacterium, revealed a mixed competitive and uncompetitive mechanism. The pH-dependence of the inhibition constants, investigated in the 6.5-8.0 range, reveals a predominant uncompetitive mechanism that increases by increasing the pH, and a lesser competitive inhibition that increases by lowering the pH. Ten crystal structures of the enzyme were independently determined using five crystals of the <scene name='59/596313/Cv/13'>native form</scene> and five crystals of the protein crystallised in the presence of fluoride. The analysis of these structures revealed the presence of <scene name='59/596313/Cv/14'>two fluoride anions coordinated to the Ni(II) ions in the active site</scene>, in terminal and bridging positions (<span style="color:gold;background-color:black;font-weight:bold;">both fluorides are colored in gold</span>). <scene name='59/596313/Cv/ | + | A strategy to control the activity of urease for medical and agricultural applications is to use enzyme inhibitors. Fluoride is a known urease inhibitor, but the structural basis of its mode of inhibition are still undetermined. Here, kinetic studies on the fluoride-induced inhibition of urease from ''Sporosarcina pasteurii'', a widespread and highly ureolytic soil bacterium, revealed a mixed competitive and uncompetitive mechanism. The pH-dependence of the inhibition constants, investigated in the 6.5-8.0 range, reveals a predominant uncompetitive mechanism that increases by increasing the pH, and a lesser competitive inhibition that increases by lowering the pH. Ten crystal structures of the enzyme were independently determined using five crystals of the <scene name='59/596313/Cv/13'>native form</scene> and five crystals of the protein crystallised in the presence of fluoride. The analysis of these structures revealed the presence of <scene name='59/596313/Cv/14'>two fluoride anions coordinated to the Ni(II) ions in the active site</scene>, in terminal and bridging positions (<span style="color:gold;background-color:black;font-weight:bold;">both fluorides are colored in gold</span>). <scene name='59/596313/Cv/20'>Click here to see animation</scene>. |
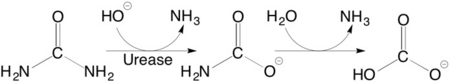
Structural studies on ureases have revealed that the immediate environment around the two Ni(II) ions at the active site is conserved, as to induce a common mechanism of catalysis whose key step is the nucleophilic attack of the nickel-bridging hydroxide on the urea molecule bound to the bimetallic nickel cluster via O and N atoms (see static image below). | Structural studies on ureases have revealed that the immediate environment around the two Ni(II) ions at the active site is conserved, as to induce a common mechanism of catalysis whose key step is the nucleophilic attack of the nickel-bridging hydroxide on the urea molecule bound to the bimetallic nickel cluster via O and N atoms (see static image below). | ||
Revision as of 13:23, 4 August 2014
| |||||||||||
- ↑ REF
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.