Sandbox bcce04
From Proteopedia
| Line 13: | Line 13: | ||
=='''Structure'''== | =='''Structure'''== | ||
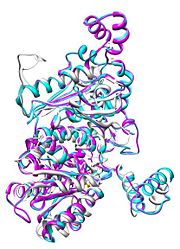
| - | [[Image:Align.jpg|thumb|left|'''Figure 1.''' Multiple alignment | + | [[Image:Align.jpg|thumb|left|'''Figure 1.''' Multiple alignment IGI - ''Geobacillus stearothermophilus'' (white),'' Homo sapiens'' (pink), ''Oryctolagus cuniculus'' (blue)<ref> Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25(13):1605-12. </ref>]] |
| - | + | Imaginary isomerase exists in the cell usually as a <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Dimer/1'>homodimer</scene>, nevertheless outside of the cell, it has been isolated as a <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Monomer/1'>monomeric</scene> structure. IGI has essentially an identical fold in all of the characterized species (see '''Figure 1'''). The <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Sec_struct/1'>secondary structure</scene> of imaginary isomerase is charaterized by an αβα conformation, on each of its two domains. The smaller domain is characterized by 5 parallel β-sheets, while the larger domain if formed out of 6 parallel/antiparallel β-sheets. Furthermore, another characteristic trait is a residue extension at the C-terminus, which wraps around the other monomer in the dimeric conformation. A "hook" that can potentially be involved in the previously mentioned extracellular activities. | |
| - | + | Imaginary isomerase has a monomer molecular mass of proximately 55 kDa. | |
'''Active Site''' - Mammalian PGI shows a degree of <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Conservation2/1'>conservation</scene> ( dark red for highly conserved regions - dark blue for variable reigions) of about 90 %. The <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Active_site2/1'>active site</scene> is the region with highest observed conservation, containing a number of residues that are crucial in the enzyme-substrate interaction mechanism (Lys210, Gln353, Glu357, Gln511, Lys518, His388b). Lys518(His388) and Glu357 <scene name='Gilman_sandbox_1/Lys_518_and_glu_357/1'>Lys 518(His388) and Glu357 </scene> are the main components of ring opening, while many of the other residues can be used for stabliziation and orientation. | '''Active Site''' - Mammalian PGI shows a degree of <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Conservation2/1'>conservation</scene> ( dark red for highly conserved regions - dark blue for variable reigions) of about 90 %. The <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Active_site2/1'>active site</scene> is the region with highest observed conservation, containing a number of residues that are crucial in the enzyme-substrate interaction mechanism (Lys210, Gln353, Glu357, Gln511, Lys518, His388b). Lys518(His388) and Glu357 <scene name='Gilman_sandbox_1/Lys_518_and_glu_357/1'>Lys 518(His388) and Glu357 </scene> are the main components of ring opening, while many of the other residues can be used for stabliziation and orientation. | ||
Revision as of 14:21, 6 August 2014
Imaginary Protein
| |||||||||||
Mechanism
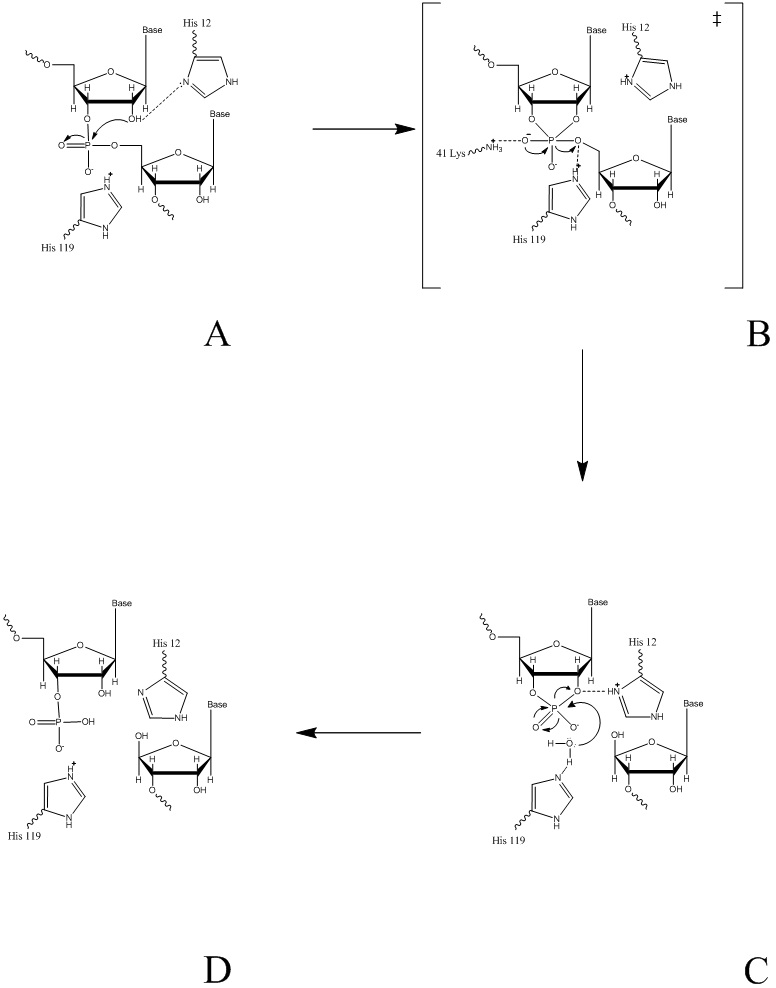
The proposed reaction mechanism of PGI for the reversible conversion of glucose-6-phosphate to fructose 6-phosphate involves an acid/base catalysis by the enzyme. The basic mechanism involves the isomerization of an aldose to a ketose. This is performed by a ring opening, followed by an isomeration of the opened ring, then a ring closing. A detailed step by step mechanism of this process can be seen as follows [8]:
Step 1. The substrate binds to the enzyme.
Step 2. The residue Lys518 (or His388b) acts as an enzymatic acid catalyzing the opening of the ring.
Step 3. Conserved Glu357 abstracts the acidic proton from C2 forming a cis-enendiol intermediate.
Step 4. Glu357 donates back the proton at the C1 position.
Step 5. Lys518 (or His388b) abstracts back the proton from the sugar ring oxygen, resulting in a ring closure, to give the product.
Regulation and Inhibition
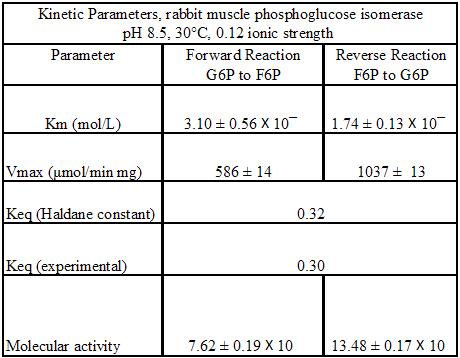
Regulation of phosphoglucoisomerase is only done by the relative concentrations of glucose-6-phosphate and fructose 6-phosphate, towards equilibrium. Nevertheless, it was found that the kinetic parameters of PGI does depend on the pH and temperature of the environment. The following kinetic parameters are proposed for rabbit PGI at pH 8.5 and 30°C [9]
It is interesting to point the regulation of PGI in other aspects that are not involved in metabolism. For example, PGI acts as a "cytokine" outside the cell in that it can be used as a cell signalling protein. PGI has been found to to be associated with AMF cells, which is found to regulate tumor cell motility. Regulation of these extracellular "cytokine" PGI/AMF can be seen. The amount of PGI/AMF that is secreted inside and outside the cell based on infection [10].
Inhibition of the phosphoglucoisomerase regulated reaction of glucose-6-phosphate to fructose-6-phosphate can also occur. Competitive competition can take place from inhibitors such as 5PAH. 5PAH resembles PGI, differing only in a nitrogen atom at the first carbon position. 5PAH is reported to have a Ki of .0000002 M [11].
Links
- Crystal Structure of human phosphoglucose isomerase (PDB=1iat)
3D structures of phosphoglucose isomerase
PGI
2pgi, 1b0z – GsPGI – Geobacillus stearothermophilus
1dqr, 1hm5, 1n8t – rPGI – rabbit
1iat, 1jlh – hPGI – human
1qxj, 1x8e, 3sxw – PfPGI – Pyrococcus furiosus
1j3p, 1j3q – TlPGI – Thermococcus litoralis
1q50 – LmPGI – Leishmania Mexicana
1u0e, 2cvp – mPGI – mouse
2q8n – PGI – Thermotoga maritima
3hjb – PGI – Vibrio cholera
3ifs – PGI – Bacillus anthracis
2wu8 – PGI – Mycobacterium tuberculosis
3ljk – FtPGI (mutant) – Francisella tularensis
3nbu – PGI – Escherichia coli
PGI complex with fructose-6-phosphate
1hox – rPGI + fructose-6-phosphate
1t10 - LmPGI + fructose-6-phosphate
2gc2 - PfPGI + fructose-6-phosphate
2cxs, 2cxt - mPGI + fructose-6-phosphate
3m5p - FtPGI (mutant) + fructose-6-phosphate
PGI complex with sorbitol-6-phosphate
1xtb - rPGI + sorbitol-6-phosphate
2gc1 - PfPGI + sorbitol-6-phosphate
2cxq - mPGI + sorbitol-6-phosphate
PGI complex with phosphoarabinose
1gzd, 1gzv – PGI + phosphoarabinose – pig
1c7r - GsPGI + phosphoarabinose
2cxp – mPGI + phosphoarabinose
1nuh - hPGI + phosphoarabinose
1qsr, 1x7n, 1x82 - PfPGI + phosphoarabinose
2gc0 - PfPGI + phosphoarabinose derivative
1koj – rPGI + phosphoarabinose derivative
PGI complex with gluconate-6-phosphate
1qy4 - PfPGI + gluconate-6-phosphate
1j3r - TlPGI + gluconate-6-phosphate
2cxr - mPGI + gluconate-6-phosphate
3q7i - FtPGI (mutant) + gluconate-6-phosphate
PGI complex with glucose-6-phosphate
1u0f - mPGI + glucose-6-phosphate
3ff1 - PGI + glucose-6-phosphate – Staphylococcus aureus
2o2c - TbPGI + glucose-6-phosphate – Trypanosoma brucei
Other PGI binary complexes
2o2d – TbPGI + citrate
1u0g, 2cxo - mPGI + erythrose-4-phosphate
3q88 – FtPGI (mutant) + ribose bisphosphate
1c7q – GsPGI + phosphate inhibitor
2cxn, 2cxu – mPGI + phosphate
1g98 – rPGI + transition state analog
2gc3 - PfPGI + mannose-6-phosphate
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ 1
- ↑ 2
- ↑ 3
- ↑ 4
- ↑ 5
- ↑ 5
- ↑ Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25(13):1605-12.
- ↑ Voet D, Voet J, and Pratt C. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. Print.
- ↑ Dyson JE, Noltmann EA. The effect of pH and temperature on the kinetic parameters of phosphoglucose isomerase. Participation of histidine and lysine in a proposed dual function mechanism. J Biol Chem. 1968 Apr 10;243(7):1401-14. PMID:5647261
- ↑ Funasaka T, Hu H, Yanagawa T, Hogan V, Raz A. Down-Regulation of Phosphoglucose Isomerase/Autocrine Motility Factors Results in Mesenchymal-to-Epithelial Transition of Human Lung Fibrosarcoma Cells. (2007) Cancer Res, 76(9)
- ↑ Arsenieva D, Hardre R, Salmon L, Jeffery CJ. The crystal structure of rabbit phosphoglucose isomerase complex with 5-phospho-D-arabinonohydroxamic acid. (2002),PNAS, 99(9)
- ↑ Lee JH, Chang KZ, Patel V, Jeffery CJ. Crystal structure of rabbit phosphoglucose isomerase complexed with its substrate D-fructose 6-phosphate. Biochemistry. 2001 Jul 3;40(26):7799-805. PMID:11425306