We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox bcce7
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
== Structural highlights == | == Structural highlights == | ||
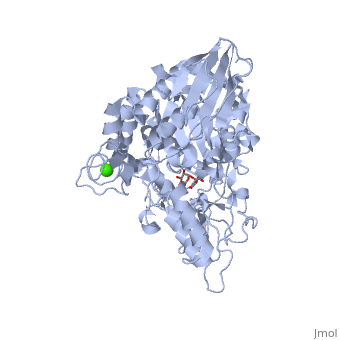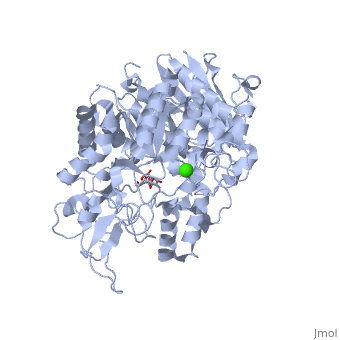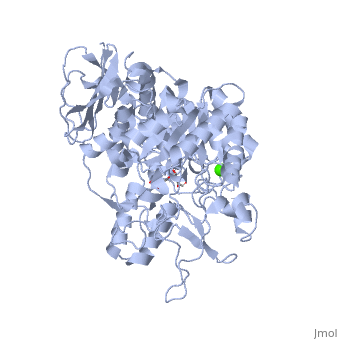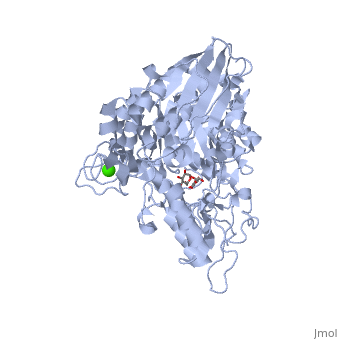
| - | This shows a broad view of the <scene name='59/596449/Active_site_broad/2'>sugar binding site</scene>. | + | This shows a broad view of the <scene name='59/596449/Active_site_broad/2'>sugar binding site</scene>. |
| + | |||
| + | Now, let's look closer at the <scene name='59/596449/Active_site_residues/1'>protein contacts with the glucose molecule</scene>. | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 14:56, 6 August 2014
S. cerevisea α-glucosidase
| |||||||||||
References
- ↑ doi: https://dx.doi.org/10.2210/pdb3axh/pdb
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644