This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
P53
From Proteopedia
| Line 43: | Line 43: | ||
'''p53 Pathway and Mutation''' | '''p53 Pathway and Mutation''' | ||
| + | In a normal cell, p53 is inactivated by its negative regulatory mdm2 (hdm2 in humans) and it is found at low levels. When DNA damage is sensed, p53's level rises. p53 binds to many regulatory sites in the genome and begins production of proteins that stop cell division until the damage is repaired. If the damage is irreparable, p53 initiates the process called programmed cell death, apoptosis, permanently removing the damage. | ||
| - | In a normal cell, p53 is inactivated by its negative regulatory mdm2 (hdm2 in humans) and it is found at low levels. When DNA damage is sensed, p53's level rises. p53 binds to many regulatory sites in the genome and begins production of proteins that stop cell division until the damage is repaired. If the damage is irreparable, p53 initiates the process called programmed cell death, apoptosis, permanently removing the damage. | ||
| - | |||
| - | |||
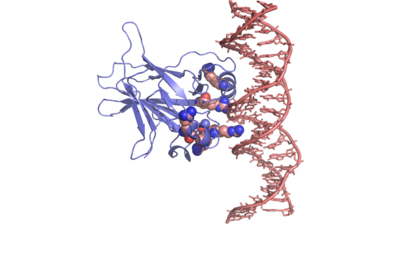
In most cases of human cancer, p53 mutations have been observed. Most of the p53 mutations that may result in cancer are found in and around the DNA-binding surface of the protein. The most common mutation changes R248, an amino acid that interacts with DNA. When mutated to another amino acid, this interaction is lost. Other residues associated with cancer-causing mutations are arginine 175, 249, 273, 282 and glycine 245. The figure at the right shows interaction of the DNA-binding domain with DNA. Key residues associated with mutations are represented by spheres. | In most cases of human cancer, p53 mutations have been observed. Most of the p53 mutations that may result in cancer are found in and around the DNA-binding surface of the protein. The most common mutation changes R248, an amino acid that interacts with DNA. When mutated to another amino acid, this interaction is lost. Other residues associated with cancer-causing mutations are arginine 175, 249, 273, 282 and glycine 245. The figure at the right shows interaction of the DNA-binding domain with DNA. Key residues associated with mutations are represented by spheres. | ||
| Line 53: | Line 51: | ||
[[Image:P53_surface_charge.png | left | 400 px | thumb]] | [[Image:P53_surface_charge.png | left | 400 px | thumb]] | ||
{{Clear}} | {{Clear}} | ||
| - | |||
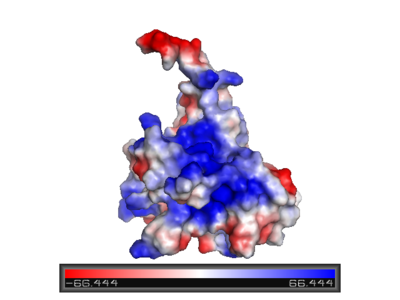
'''Surface charge of the DNA binding domain''' | '''Surface charge of the DNA binding domain''' | ||
The figure at the left shows the surface charge of the p53 DNA-binding domain. It is rich in arginine amino acids that interact with DNA, and this causes its surface to be positively charged. This domain recognizes specific regulatory sites on the DNA. The flexible structure of p53 allows it to bind to many different variants of binding sites, allowing it to regulate transcription at many places in the genome. | The figure at the left shows the surface charge of the p53 DNA-binding domain. It is rich in arginine amino acids that interact with DNA, and this causes its surface to be positively charged. This domain recognizes specific regulatory sites on the DNA. The flexible structure of p53 allows it to bind to many different variants of binding sites, allowing it to regulate transcription at many places in the genome. | ||
| - | |||
| - | |||
| - | |||
---- | ---- | ||
| - | |||
| - | |||
| - | |||
| - | |||
<scene name='Sandbox/P53_dna_binding_domain/1'>Click Here to view a Three-dimensional Representation of the DNA-binding Domain Bound to DNA</scene> | <scene name='Sandbox/P53_dna_binding_domain/1'>Click Here to view a Three-dimensional Representation of the DNA-binding Domain Bound to DNA</scene> | ||
| - | |||
There is a Zn-binding motif on p53. The p53 Zn atom is coordinated by residues | There is a Zn-binding motif on p53. The p53 Zn atom is coordinated by residues | ||
| Line 76: | Line 65: | ||
coordination. The Zn has been represented as a red sphere in the figure at the right. | coordination. The Zn has been represented as a red sphere in the figure at the right. | ||
</StructureSection> | </StructureSection> | ||
| - | __NOTOC__ | ||
==3D structures of p53== | ==3D structures of p53== | ||
Revision as of 09:35, 19 August 2014
| |||||||||||
Contents |
3D structures of p53
Updated on 19-August-2014
p53 DNA-binding domain
2xwr, 2ocj, 2ybg – h-p53 DBD – human
2fej - h-p53 DBD - NMR
2wgx, 3d05, 3d06, 3d07, 3d08, 3d09, 2qvq, 2qxa, 2qxb, 2qxc, 2pcx, 2j1w, 2j1x, 2j1y, 2j1z, 2j20, 2j21, 2bim, 2bin, 2bio, 2bip, 2biq, 1uol, 4kvp, 4lo9, 4loe, 4lof, 4ibq, 4ibs, 4ibt, 4iby, 4ibz, 4ijt, 4mzi - h-p53 DBD (mutant)
1hs5 - h-p53 residues 324-357 - NMR
3q01 - h-p53 DBD + TD
2ioi, 2ioo, 1hu8 – m-p53 DBD – mouse
1t4w – p53-like DBD – Caenoharbditis elegans
p53 DNA-binding domain complex with DNA
3igk, 3igl, 3kz8, 3kmd, 2ac0, 2ady, 2ahi, 2ata, 1tsr, 1tup, 4hje – h-p53 DBD + DNA
3d0a, 3ts8, 4ibu, 4ibv, 4ibw - h-p53 DBD (mutant) + DNA
3q05, 3q06 - h-p53 DBD + TD + DNA
3exj, 3exl, 2p52, 2geq – m-p53 DBD + DNA
p53 DNA-binding domain complex with small molecule
2x0u, 2x0v, 2x0w, 4agl, 4agm, 4agn, 4ago, 4agp, 4agq – h-p53 DBD (mutant) + benzene derivative
3zme - h-p53 DBD (mutant) + pyrazol derivative
2vuk - h-p53 DBD + drug
2iom - m-p53 DBD + propanol
p53 DNA-binding domain complex with protein
2k8f – h-p53 residues 1-39 + histone acetyltransferase (mutant) – NMR
2h1l - h-p53 DBD + large T antigen
1ycs - h-p53 DBD + 53BP2
1gzh, 1kzy - h-p53 DBD + tumor suppressor p53-binding protein
p53 transactivation domain
2z5s, 2z5t - h-p53 TAD + MDM4 protein
1ycq, 1ycr - h-p53 TAD + MDM2 protein
2l14 – h-p53 TAD + CREB-binding protein
2gs0 – h-p53 TAD + RNA polymerase II transcription factor
p53 tetramerization domain
2j0z, 3sak, 1sae, 1saf, 1sah, 1saj, 1sak, 1sal, 1olh, 1pes, 1pet, 1olg – h-p53 TD – NMR
1c26, 1aie - h-p53 TD
2j10, 2j11, 1a1u - h-p53 TD (mutant) – NMR
p53 tetramerization+DBD domains
4mzr – h-p53 + DNA
Additional Resources
For additional information, see: Cancer
For additional information, see: Oncogenes
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Michal Harel, Eran Hodis, Mary Ball, Alexander Berchansky, David Canner