This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Pepsin
From Proteopedia
| Line 20: | Line 20: | ||
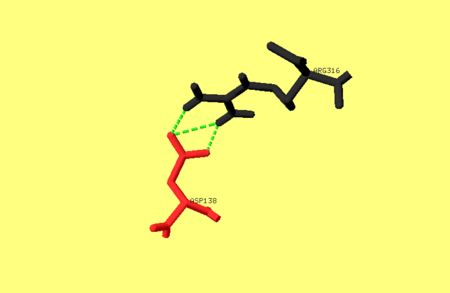
One of the main salt bridges in pepsin includes residues from one of the dyad-related hylices (h’N and h’C) and a residue from the opposite lobe, e.g. Asp138 interacts with Arg316. For an image of these residues refer to image 1. bellow. This ion-pair may be important for the stability of the pepsin fold, since chemical modification of Arg316 with butane-2,3-dione partially inactivates the enzyme, as does treatment with phenylglyoxal <ref name="Xray" />. | One of the main salt bridges in pepsin includes residues from one of the dyad-related hylices (h’N and h’C) and a residue from the opposite lobe, e.g. Asp138 interacts with Arg316. For an image of these residues refer to image 1. bellow. This ion-pair may be important for the stability of the pepsin fold, since chemical modification of Arg316 with butane-2,3-dione partially inactivates the enzyme, as does treatment with phenylglyoxal <ref name="Xray" />. | ||
Image 1. The hydrogen bonds between Asp138 and Arg316[[Image:Hydrogen bond.png|450 px ]] | Image 1. The hydrogen bonds between Asp138 and Arg316[[Image:Hydrogen bond.png|450 px ]] | ||
| - | |||
There are many sub domains in pepsin. One sub domain which includes the residues 222-235; 255-276; and 283-290. Portions of this sub domains can interact with inhibitors, and therefore contribute to the structural delineation of S’ sub sites <ref name="flexible" />. It is also possible that ridged movements of sub domains relative to the large domain may help the enzyme adjust to various substrate structures. Furthermore, local regional flexibility of structures such as the “flap,” in and around the active site of pepsin has been suggested to modulate substrate and inhibitor binding <ref name="flexible" />. | There are many sub domains in pepsin. One sub domain which includes the residues 222-235; 255-276; and 283-290. Portions of this sub domains can interact with inhibitors, and therefore contribute to the structural delineation of S’ sub sites <ref name="flexible" />. It is also possible that ridged movements of sub domains relative to the large domain may help the enzyme adjust to various substrate structures. Furthermore, local regional flexibility of structures such as the “flap,” in and around the active site of pepsin has been suggested to modulate substrate and inhibitor binding <ref name="flexible" />. | ||
| Line 26: | Line 25: | ||
Once denatured, pepsin is unable to refold to an active native state upon returning from denaturing conditions. One proposed solution for this is that pepsin formation depends on a separate prosegment (PS) domain. When returning from the denatured state, the denatured pepsin first has to bypass a large folding barrier and then in the presence of PS the native state can become thermodynamically stable. The PS therefore can catalyze pepsin folding by stabilizing the transition state <ref name="native" /> . | Once denatured, pepsin is unable to refold to an active native state upon returning from denaturing conditions. One proposed solution for this is that pepsin formation depends on a separate prosegment (PS) domain. When returning from the denatured state, the denatured pepsin first has to bypass a large folding barrier and then in the presence of PS the native state can become thermodynamically stable. The PS therefore can catalyze pepsin folding by stabilizing the transition state <ref name="native" /> . | ||
</StructureSection> | </StructureSection> | ||
| - | __NOTOC__ | ||
==3D structures of pepsin== | ==3D structures of pepsin== | ||
Revision as of 09:57, 19 August 2014
| |||||||||||
3D structures of pepsin
Updated on 19-August-2014
Pepsin
2h6s, 2qzw – CaPep residues 59-398 – Candida albicans
1s2b – SlPep - Scytalidium lignicola
1y43 – Pep – Aspergillus niger
1ibq - Pep – Aspergillus phoenicis
5pep, 3pep, 4pep – pPep – pig
1uh7, 1uh8, 1uh9, 2apr – RcPep – Rhizopus chinensis
1mpp – Pep – Rhizomucor pusillus
1fmu, 1fmx, 2jxr – yPep – yeast
1flh, 1psn, 3utl – hPep – human
3app – PjPep – Penicillium janthinellum
1am5 – Pep – Atlantic cod
1er8, 3er3, 4er1, 4ape, 1ent – CpPep – Cryphonectria parasitica
3lzy, 1oew – CpPep residues 90-419
Pepsinogen
3psg, 2psg - pPepn
1avf - hPepn
Pepsin complex with inhibitor
2h6t - CaPep residues 59-398 + pepstatin
1eag - CaPep residues 59-398 + inhibitor
2qzx - CaPep residues 77-418 + pepstatin
1j71 - Pep + peptide – Candida tropicalis<br />
2ifr, 2ifw, 1s2k – SlPep + peptide
1yx9 – pPep + DMSO
1psa, 1f34 – pPep + inhibitor
1wkr - Pep + pepstatin – Irpex lacteus
4apr, 5apr, 6apr - RcPep + pepstatin
3apr - RcPep + peptide
2rmp - Pep + pepstatin – Rhizomucor miehei
1nlu, 1ga1, 1ga4 - PsPep + pseudo-iodotyrostatin – Pseudomonas
1kdv, 1kdy – PsPep + peptide
1kdz, 1ke1, 1ke2, 1ga6 - PsPep + statin inhibitor
1g0v – yPep + protease A inhibitor
1fq4, 1fq5, 1fq6, 1fq7, 1fq8 - yPep + inhibitor
1qrp – hPep + peptide
1pso – hPep + pepstatin
1bxo, 1bxq, 2wea, 2web, 2wec, 2wed - PjPep + inhibitor
1ppk, 1apv, 1apw, 1ppl, 1ppm – PjPep + peptide analog
1apt, 1apu – PjPep + pepstatin
Endothiapepsin
1er8, 3er3, 4er1, 4ape, 1ent – CpPep – Cryphonectria parasitica
3lzy, 1oew – CpPep residues 90-419
3urj - ETPep – Endothia parasitica
3pb5, 3pbd, 3pbz, 3pcw, 3pgi, 3pi0, 3pld, 3pll, 3pm4, 3pmu, 3pmy – CpETPep + fragment chemotype
3pcz – CpETPep + benzamidine
3prs, 3pww – CpETPep + antiviral drug
3q6y, 1gvx, 1gvu, 1gvv, 1gvw, 1gvt, 2jji, 2jjj - CpETPep + pyrrolidine derivative
3t6i - CpETPep + azepine derivative
3t7p, 4lhh, 4lbt, 4kup - CpETPep + hydrazine derivative
4l6b, 3t7x, 3t7q, 3psy – CpETPwo + thiophen inhibitor
3er5, 4er4, 5er2, 2er0, 2er6, 2er7, 2er9, 5er1, 1epl, 1epm, 1epn, 1epo, 1epp, 1epq, 1epr, 3uri, 3url – CpETPep + peptide
4er2 - CpETPep + pepstatin
2v00, 1od1, 1oex, 1eed, 1e81, 1e82, 1e80, 1e5o – CpETPep + inhibitor
2vs2, 1gkt - CpETPep + inhibitor - neutron
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Cooper JB, Khan G, Taylor G, Tickle IJ, Blundell TL. X-ray analyses of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 A resolution. J Mol Biol. 1990 Jul 5;214(1):199-222. PMID:2115088
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Abad-Zapatero C, Rydel TJ, Erickson J. Revised 2.3 A structure of porcine pepsin: evidence for a flexible subdomain. Proteins. 1990;8(1):62-81. PMID:2217165 doi:http://dx.doi.org/10.1002/prot.340080109
- ↑ 3.0 3.1 The prosegment catalyzed pepsin folding to a kinetically trapped native state. Biochemistry 49:365-371
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky