This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2pea
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
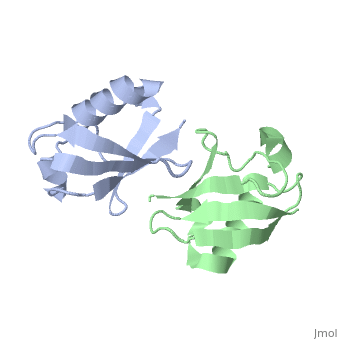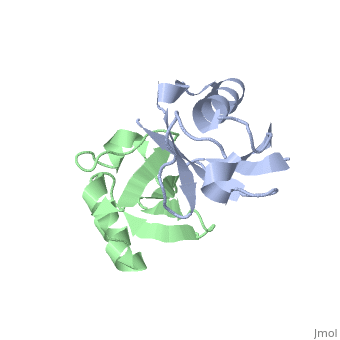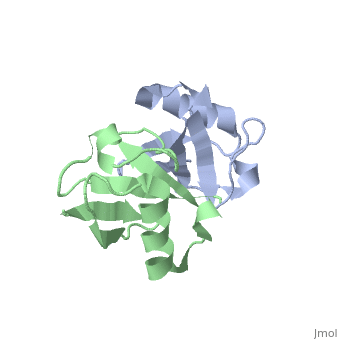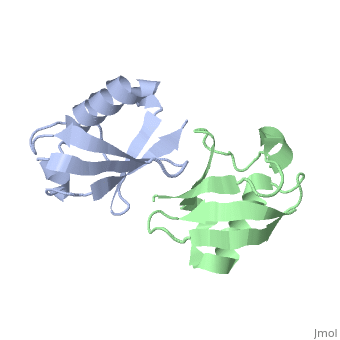
| - | [[ | + | ==NMR Based Structure of the Closed Conformation of LYS48-Linked Di-Ubiquitin Using Experimental Global Rotational Diffusion Tensor from NMR Relaxation Measurements== |
| + | <StructureSection load='2pea' size='340' side='right' caption='[[2pea]], [[NMR_Ensembles_of_Models | 1 NMR models]]' scene=''> | ||
| + | == Structural highlights == | ||
| + | <table><tr><td colspan='2'>[[2pea]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2PEA OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2PEA FirstGlance]. <br> | ||
| + | </td></tr><tr><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1d3z|1d3z]], [[1aar|1aar]], [[2bgf|2bgf]], [[2pe9|2pe9]]</td></tr> | ||
| + | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2pea FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2pea OCA], [http://www.rcsb.org/pdb/explore.do?structureId=2pea RCSB], [http://www.ebi.ac.uk/pdbsum/2pea PDBsum]</span></td></tr> | ||
| + | <table> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/pe/2pea_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | We present a simple and robust approach that uses the overall rotational diffusion tensor as a structural constraint for domain positioning in multidomain proteins and protein-protein complexes. This method offers the possibility to use NMR relaxation data for detailed structure characterization of such systems provided the structures of individual domains are available. The proposed approach extends the concept of using long-range information contained in the overall rotational diffusion tensor. In contrast to the existing approaches, we use both the principal axes and principal values of protein's rotational diffusion tensor to determine not only the orientation but also the relative positioning of the individual domains in a protein. This is achieved by finding the domain arrangement in a molecule that provides the best possible agreement with all components of the overall rotational diffusion tensor derived from experimental data. The accuracy of the proposed approach is demonstrated for two protein systems with known domain arrangement and parameters of the overall tumbling: the HIV-1 protease homodimer and Maltose Binding Protein. The accuracy of the method and its sensitivity to domain positioning are also tested using computer-generated data for three protein complexes, for which the experimental diffusion tensors are not available. In addition, the proposed method is applied here to determine, for the first time, the structure of both open and closed conformations of a Lys48-linked diubiquitin chain, where domain motions render impossible accurate structure determination by other methods. The proposed method opens new avenues for improving structure characterization of proteins in solution. | ||
| - | + | Structural assembly of multidomain proteins and protein complexes guided by the overall rotational diffusion tensor.,Ryabov Y, Fushman D J Am Chem Soc. 2007 Jun 27;129(25):7894-902. Epub 2007 Jun 6. PMID:17550252<ref>PMID:17550252</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==See Also== | ==See Also== | ||
*[[Ubiquitin|Ubiquitin]] | *[[Ubiquitin|Ubiquitin]] | ||
| - | + | == References == | |
| - | == | + | <references/> |
| - | < | + | __TOC__ |
| + | </StructureSection> | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Fushman, D.]] | [[Category: Fushman, D.]] | ||
Revision as of 06:30, 29 September 2014
NMR Based Structure of the Closed Conformation of LYS48-Linked Di-Ubiquitin Using Experimental Global Rotational Diffusion Tensor from NMR Relaxation Measurements
| |||||||||||