This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
1cfc
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | ==CALCIUM-FREE CALMODULIN== | |
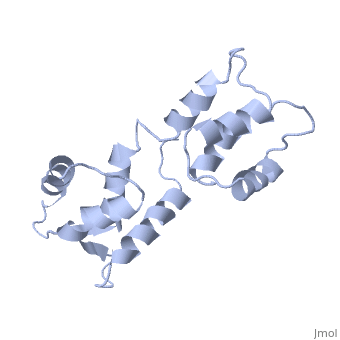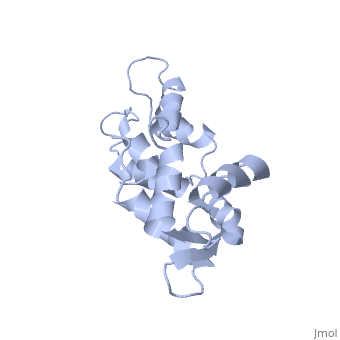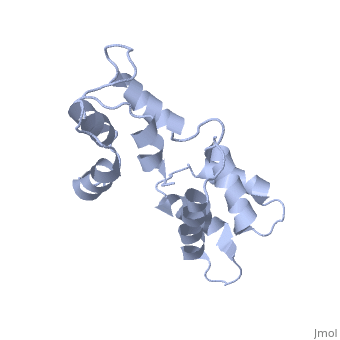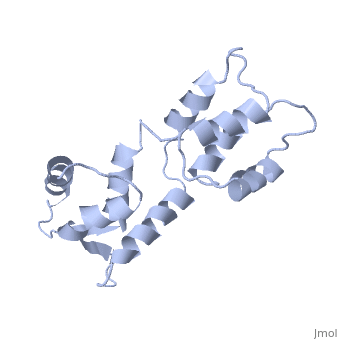
| - | === | + | <StructureSection load='1cfc' size='340' side='right' caption='[[1cfc]], [[NMR_Ensembles_of_Models | 25 NMR models]]' scene=''> |
| - | + | == Structural highlights == | |
| + | <table><tr><td colspan='2'>[[1cfc]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Xenopus_laevis Xenopus laevis]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1CFC OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1CFC FirstGlance]. <br> | ||
| + | </td></tr><tr><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1cfd|1cfd]]</td></tr> | ||
| + | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1cfc FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1cfc OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1cfc RCSB], [http://www.ebi.ac.uk/pdbsum/1cfc PDBsum]</span></td></tr> | ||
| + | <table> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/cf/1cfc_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | The three-dimensional structure of calmodulin in the absence of Ca2+ has been determined by three- and four-dimensional heteronuclear NMR experiments, including ROE, isotope-filtering combined with reverse labelling, and measurement of more than 700 three-bond J-couplings. In analogy with the Ca(2+)-ligated state of this protein, it consists of two small globular domains separated by a flexible linker, with no stable, direct contacts between the two domains. In the absence of Ca2+, the four helices in each of the two globular domains form a highly twisted bundle, capped by a short anti-parallel beta-sheet. This arrangement is qualitatively similar to that observed in the crystal structure of the Ca(2+)-free N-terminal domain of troponin C. | ||
| - | + | Solution structure of calcium-free calmodulin.,Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A Nat Struct Biol. 1995 Sep;2(9):768-76. PMID:7552748<ref>PMID:7552748</ref> | |
| - | + | ||
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
==See Also== | ==See Also== | ||
| Line 11: | Line 30: | ||
*[[NMR Ensembles of Models|NMR Ensembles of Models]] | *[[NMR Ensembles of Models|NMR Ensembles of Models]] | ||
*[[Structural alignment tools|Structural alignment tools]] | *[[Structural alignment tools|Structural alignment tools]] | ||
| - | + | == References == | |
| - | == | + | <references/> |
| - | < | + | __TOC__ |
| + | </StructureSection> | ||
[[Category: Xenopus laevis]] | [[Category: Xenopus laevis]] | ||
[[Category: Bax, A.]] | [[Category: Bax, A.]] | ||
Revision as of 13:57, 29 September 2014
CALCIUM-FREE CALMODULIN
| |||||||||||