We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
2gr9
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
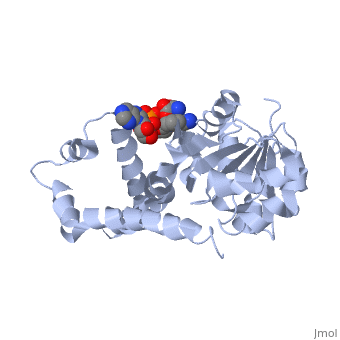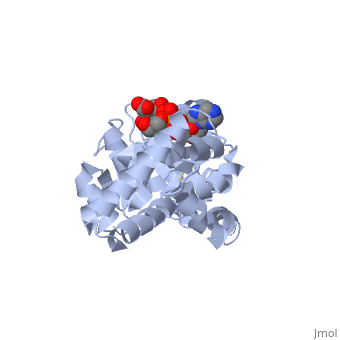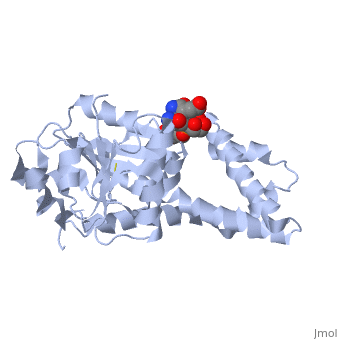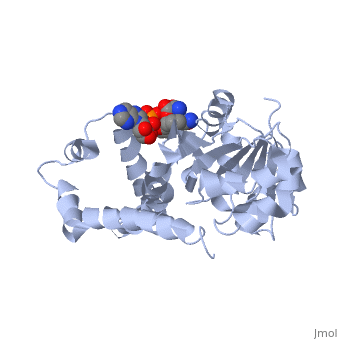
| - | + | ==Crystal structure of P5CR complexed with NADH== | |
| - | === | + | <StructureSection load='2gr9' size='340' side='right' caption='[[2gr9]], [[Resolution|resolution]] 3.10Å' scene=''> |
| - | + | == Structural highlights == | |
| - | + | <table><tr><td colspan='2'>[[2gr9]] is a 5 chain structure with sequence from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2GR9 OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2GR9 FirstGlance]. <br> | |
| - | ==Disease== | + | </td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=GLU:GLUTAMIC+ACID'>GLU</scene>, <scene name='pdbligand=NAI:1,4-DIHYDRONICOTINAMIDE+ADENINE+DINUCLEOTIDE'>NAI</scene><br> |
| + | <tr><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[2gra|2gra]]</td></tr> | ||
| + | <tr><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Pyrroline-5-carboxylate_reductase Pyrroline-5-carboxylate reductase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.5.1.2 1.5.1.2] </span></td></tr> | ||
| + | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2gr9 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2gr9 OCA], [http://www.rcsb.org/pdb/explore.do?structureId=2gr9 RCSB], [http://www.ebi.ac.uk/pdbsum/2gr9 PDBsum]</span></td></tr> | ||
| + | <table> | ||
| + | == Disease == | ||
[[http://www.uniprot.org/uniprot/P5CR1_HUMAN P5CR1_HUMAN]] Defects in PYCR1 are the cause of cutis laxa autosomal recessive type 2B (ARCL2B) [MIM:[http://omim.org/entry/612940 612940]]. A multisystem disorder characterized by the appearance of premature aging, wrinkled and lax skin with reduced elasticity, joint laxity, craniofacial dysmorphic features, intrauterine growth retardation with some degree of postnatal growth deficiency, and developmental delay.<ref>PMID:19648921</ref> <ref>PMID:19576563</ref> Defects in PYCR1 are the cause of cutis laxa, autosomal recessive, type 3B (ARCL3B) [MIM:[http://omim.org/entry/614438 614438]]. ARCL3B is a disorder characterized by an aged appearance with distinctive facial features, sparse hair, ophthalmologic abnormalities, intrauterine growth retardation, and cutis laxa.<ref>PMID:19648921</ref> <ref>PMID:22052856</ref> | [[http://www.uniprot.org/uniprot/P5CR1_HUMAN P5CR1_HUMAN]] Defects in PYCR1 are the cause of cutis laxa autosomal recessive type 2B (ARCL2B) [MIM:[http://omim.org/entry/612940 612940]]. A multisystem disorder characterized by the appearance of premature aging, wrinkled and lax skin with reduced elasticity, joint laxity, craniofacial dysmorphic features, intrauterine growth retardation with some degree of postnatal growth deficiency, and developmental delay.<ref>PMID:19648921</ref> <ref>PMID:19576563</ref> Defects in PYCR1 are the cause of cutis laxa, autosomal recessive, type 3B (ARCL3B) [MIM:[http://omim.org/entry/614438 614438]]. ARCL3B is a disorder characterized by an aged appearance with distinctive facial features, sparse hair, ophthalmologic abnormalities, intrauterine growth retardation, and cutis laxa.<ref>PMID:19648921</ref> <ref>PMID:22052856</ref> | ||
| - | + | == Function == | |
| - | ==Function== | + | |
[[http://www.uniprot.org/uniprot/P5CR1_HUMAN P5CR1_HUMAN]] Housekeeping enzyme that catalyzes the last step in proline biosynthesis. Can utilize both NAD and NADP, but has higher affinity for NAD. Involved in the cellular response to oxidative stress.<ref>PMID:19648921</ref> <ref>PMID:16730026</ref> | [[http://www.uniprot.org/uniprot/P5CR1_HUMAN P5CR1_HUMAN]] Housekeeping enzyme that catalyzes the last step in proline biosynthesis. Can utilize both NAD and NADP, but has higher affinity for NAD. Involved in the cellular response to oxidative stress.<ref>PMID:19648921</ref> <ref>PMID:16730026</ref> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/gr/2gr9_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Pyrroline-5-carboxylate reductase (P5CR) is a universal housekeeping enzyme that catalyzes the reduction of Delta(1)-pyrroline-5-carboxylate (P5C) to proline using NAD(P)H as the cofactor. The enzymatic cycle between P5C and proline is very important for the regulation of amino acid metabolism, intracellular redox potential, and apoptosis. Here, we present the 2.8 Angstroms resolution structure of the P5CR apo enzyme, its 3.1 Angstroms resolution ternary complex with NAD(P)H and substrate-analog. The refined structures demonstrate a decameric architecture with five homodimer subunits and ten catalytic sites arranged around a peripheral circular groove. Mutagenesis and kinetic studies reveal the pivotal roles of the dinucleotide-binding Rossmann motif and residue Glu221 in the human enzyme. Human P5CR is thermostable and the crystals were grown at 37 degrees C. The enzyme is implicated in oxidation of the anti-tumor drug thioproline. | ||
| - | + | Crystal structure of human pyrroline-5-carboxylate reductase.,Meng Z, Lou Z, Liu Z, Li M, Zhao X, Bartlam M, Rao Z J Mol Biol. 2006 Jun 23;359(5):1364-77. Epub 2006 May 11. PMID:16730026<ref>PMID:16730026</ref> | |
| - | + | ||
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
==See Also== | ==See Also== | ||
*[[Pyrroline-5-carboxylate reductase|Pyrroline-5-carboxylate reductase]] | *[[Pyrroline-5-carboxylate reductase|Pyrroline-5-carboxylate reductase]] | ||
| - | + | == References == | |
| - | == | + | <references/> |
| - | + | __TOC__ | |
| + | </StructureSection> | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Pyrroline-5-carboxylate reductase]] | [[Category: Pyrroline-5-carboxylate reductase]] | ||
Revision as of 05:13, 3 October 2014
Crystal structure of P5CR complexed with NADH
| |||||||||||