Sigma factor
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
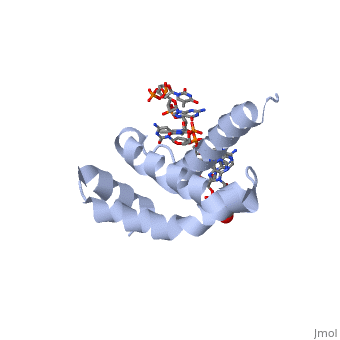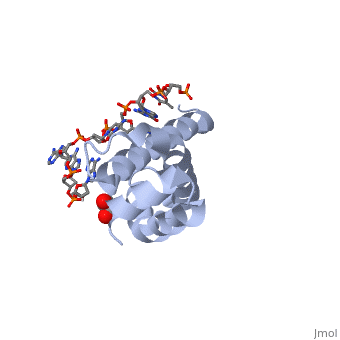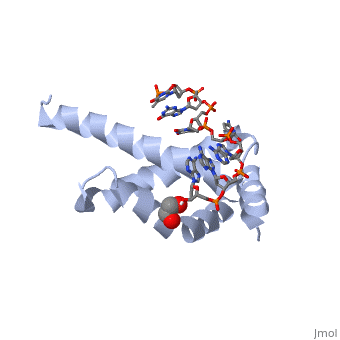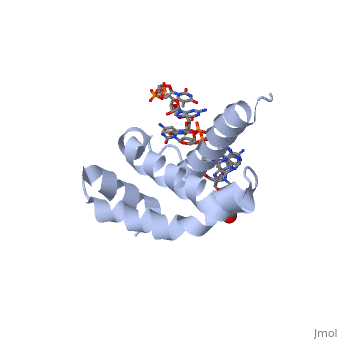
There are many types of σ-subunits, and each recognizes a unique promoter sequence. Furthmore, each unique σ is composed of a variable number of structured domains. The simplest σ-factors have two domains, few have three, and most, called '''housekeeping σ-factors''', have 4 domains, given the names σ(4), σ(3), σ(2), and σ(1.1). All domains are linked by very flexible peptide '''linkers''' which can extend very long distances. Each of these domains utilizes DNA-binding determinants, or domains that recognize specific sequences and conformations in DNA. Most commonly, these recognized sequences occur at the -35 and -10 locations upstream of the +1 site. One such DNA-binding motif, '''the helix-turn-helix motif''' (<scene name='59/591940/Hth_motif/2'>HTH</scene>), helps specifically recognize DNA promoters at both the -35 and -10 positions. This HTH motif, used by most σ-factors, maintains its specificity and accuracy by binding in the '''major groove''' of DNA, where it can interact with the base pairs in the DNA double-helix. In many prokaryotes, these portions of DNA maintain consensus adenosine and thymine sequences, such as <scene name='59/591940/Ta_sequence/1'>TATAAT</scene>. | There are many types of σ-subunits, and each recognizes a unique promoter sequence. Furthmore, each unique σ is composed of a variable number of structured domains. The simplest σ-factors have two domains, few have three, and most, called '''housekeeping σ-factors''', have 4 domains, given the names σ(4), σ(3), σ(2), and σ(1.1). All domains are linked by very flexible peptide '''linkers''' which can extend very long distances. Each of these domains utilizes DNA-binding determinants, or domains that recognize specific sequences and conformations in DNA. Most commonly, these recognized sequences occur at the -35 and -10 locations upstream of the +1 site. One such DNA-binding motif, '''the helix-turn-helix motif''' (<scene name='59/591940/Hth_motif/2'>HTH</scene>), helps specifically recognize DNA promoters at both the -35 and -10 positions. This HTH motif, used by most σ-factors, maintains its specificity and accuracy by binding in the '''major groove''' of DNA, where it can interact with the base pairs in the DNA double-helix. In many prokaryotes, these portions of DNA maintain consensus adenosine and thymine sequences, such as <scene name='59/591940/Ta_sequence/1'>TATAAT</scene>. | ||
| - | ==Restriction== | + | ===Restriction=== |
Normally, σ-factor domains cannot bind to promoters. These domains usually are placed in very compacted positions relative to each other, a conformation that buries DNA-binding determinants. This type of restriction is called '''conformational restriction'''. Additionally, in housekeeping σs, a domain called the '''σ(1.1)''' stabilizes the compact conformation mentioned above, thereby preventing any promoter recognition. | Normally, σ-factor domains cannot bind to promoters. These domains usually are placed in very compacted positions relative to each other, a conformation that buries DNA-binding determinants. This type of restriction is called '''conformational restriction'''. Additionally, in housekeeping σs, a domain called the '''σ(1.1)''' stabilizes the compact conformation mentioned above, thereby preventing any promoter recognition. | ||
Revision as of 02:22, 14 October 2014
| |||||||||||