We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Citrate Synthase
From Proteopedia
(Difference between revisions)
m |
|||
| Line 1: | Line 1: | ||
<StructureSection load='1cts' size='450' side='right' scene='User:Wayne_Decatur/1cts_to_2cts_(citrate_synthase)_morph_methods/Camorph/5' caption='Open conformation of citrate synthase dimer complex with citrate (PDB code [[1cts]]) and closed conformation of citrate synthase dimer complex with citrate and CoA (PDB code [[2cts]])'> | <StructureSection load='1cts' size='450' side='right' scene='User:Wayne_Decatur/1cts_to_2cts_(citrate_synthase)_morph_methods/Camorph/5' caption='Open conformation of citrate synthase dimer complex with citrate (PDB code [[1cts]]) and closed conformation of citrate synthase dimer complex with citrate and CoA (PDB code [[2cts]])'> | ||
| - | == | + | ==Overview== |
'''Citrate synthase''' is an enzyme active in all examined cells, where it is most often responsible for catalyzing the first reaction of the [[The Citric Acid Cycle|citric acid cycle (Krebs Cycle or the tricarboxylic acid <nowiki>[</nowiki>TCA<nowiki>]</nowiki> cycle)]]: the condensation of acetyl-CoA and oxaloacetate to form citrate. Although in eukaryotes it is a mitochondrial enzyme, and in fact, is often used as a enzyme marker for intact mitochondria, it is encoded by nuclear DNA<ref>[http://en.wikipedia.org/wiki/Citrate_synthase "Citrate Synthase -." Wikipedia, the Free Encyclopedia. Web. 22 Mar. 2010].</ref>. The standard free energy change (ΔG°’) for the citrate synthase reaction is | '''Citrate synthase''' is an enzyme active in all examined cells, where it is most often responsible for catalyzing the first reaction of the [[The Citric Acid Cycle|citric acid cycle (Krebs Cycle or the tricarboxylic acid <nowiki>[</nowiki>TCA<nowiki>]</nowiki> cycle)]]: the condensation of acetyl-CoA and oxaloacetate to form citrate. Although in eukaryotes it is a mitochondrial enzyme, and in fact, is often used as a enzyme marker for intact mitochondria, it is encoded by nuclear DNA<ref>[http://en.wikipedia.org/wiki/Citrate_synthase "Citrate Synthase -." Wikipedia, the Free Encyclopedia. Web. 22 Mar. 2010].</ref>. The standard free energy change (ΔG°’) for the citrate synthase reaction is | ||
-31.5kJ/mol <ref name="voet">Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008.</ref>. This negative free energy value means that citrate synthase is likely to function far from equilibrium under physiological conditions, and is thus a rate-determining enzyme in the citric acid cycle. | -31.5kJ/mol <ref name="voet">Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008.</ref>. This negative free energy value means that citrate synthase is likely to function far from equilibrium under physiological conditions, and is thus a rate-determining enzyme in the citric acid cycle. | ||
| + | |||
| + | ==Structure== | ||
| - | + | Biologically, citrate synthase exists as a <scene name='Daniel_Eddelman_Sandbox_2/Cts_open_monomer/2'>homodimer</scene> of a single amino acid chain <scene name='Daniel_Eddelman_Sandbox_2/Cts_open_monomer/1'>monomer</scene>. Each identical subunit consists of a large and a small domain, and is comprised almost entirely of α helices (making it an all α protein). In its free enzyme state, citrate synthase exists in <scene name='Daniel_Eddelman_Sandbox_2/Cts_open_monomer/2'>an “open” form of the homodimer</scene>, with its two domains forming a cleft containing the substrate (oxaloacetate) binding site (PDB: [[1cts]]) <ref name="1cts">PMID:7120407</ref><ref name="substrate note">In this structure [[1cts]], citrate, the resulting product of the conversion, is actually bound where oxaloacetate binds.</ref>. When oxaloacetate binds, the smaller domain undergoes an 18° rotation, sealing the oxaloacetate binding site<ref>PMID:7308213</ref> and resulting in the <scene name='Daniel_Eddelman_Sandbox_2/Closed_homodimer/1'>closed conformation of the homodimer</scene> (PDB: [[2cts]])<ref name="1cts" />. The dramatic conformational change is best illustrated via <scene name='User:Wayne_Decatur/1cts_to_2cts_(citrate_synthase)_morph_methods/Camorph/5'>a morph between the "open" and "closed" states</scene>, and be sure to view <scene name='User:Wayne_Decatur/1cts_to_2cts_(citrate_synthase)_morph_methods/Camorphside/3'>the morph from the side</scene> as well to get a full sense of the structural change. The conformational change not only prevents solvent from reaching the bound substrate, but also generates the acetyl-CoA binding site. This presence of “open” and “closed” forms results in citrate synthase having Ordered Sequential kinetic behavior <ref name="voet" />.<br> | |
{{Button Toggle AnimationOnPause}}<br> | {{Button Toggle AnimationOnPause}}<br> | ||
{{Link Toggle FancyCartoonHighQualityView}} | {{Link Toggle FancyCartoonHighQualityView}} | ||
| - | + | ==Mechanism== | |
| + | |||
| + | <scene name='Citrate_Synthase/5ctsactivesitedimer/2'>Three side chains in each of the two active sites</scene> of the dimer contribute directly to the chemistry of catalysis. Focusing on a single active site in the closed conformation, one can easily observe that <scene name='Citrate_Synthase/5ctsactivesiteresidues/10'>these three side chains and the two substrates are together</scene> in an arrangement favorable for reaction. (By contrast, <scene name='Citrate_Synthase/Activesite1ctsto2cts/12'>the active site residues are significantly farther apart</scene> in the open conformation; the difference in the distance is ~5Å along the axis that changes the most during the conformation shift.) {{Link Toggle AnimationOnPause}} | ||
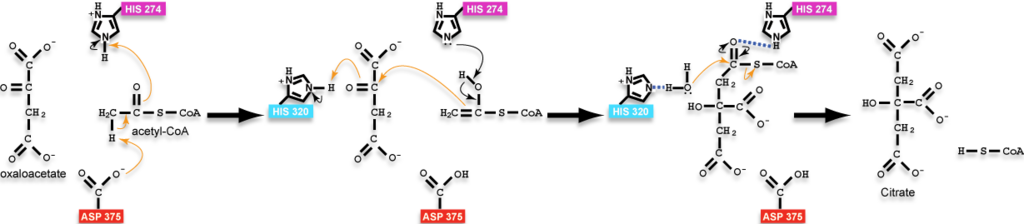
The reaction mechanism for citrate synthase was proposed by Remington and colleagues<ref name="1cts">PMID:7120407</ref><ref>PMID: 2337600</ref> and is illustrated here in three dimensions using structures resembling key states of the reaction<ref>[[5cts]] as the state preceding condensation with oxaloacetate and a non-reactive version of acetyl-CoA bound, [[6cts]] as the state containing the bound intermediate, and [[3cts]] as the complex with the products. Positions of hydrogens on the ligands were calculated and added back to structures in the reaction scheme for instructional purposes and are not present in the experimentally-determined structures; additionally, arrows are drawn with atoms of the analog of acetyl-CoA to approximate the position of the reactive groups only as the reactive groups are not actually part of the analog or the molecules would have reacted; please, see the reaction scheme on this page for a more thorough accounting of the chemistry.</ref>. In this mechanism, three ionizable side chains in the active site of citrate synthase participate in acid-base catalysis: <scene name='Citrate_Synthase/5ctsactivesiteresidues/10'>His 274, His 320, and Asp 375</scene>. Citrate synthase is among one of the few enzymes that can directly form a carbon-carbon bond without the presence of metal ion cofactors.<br> | The reaction mechanism for citrate synthase was proposed by Remington and colleagues<ref name="1cts">PMID:7120407</ref><ref>PMID: 2337600</ref> and is illustrated here in three dimensions using structures resembling key states of the reaction<ref>[[5cts]] as the state preceding condensation with oxaloacetate and a non-reactive version of acetyl-CoA bound, [[6cts]] as the state containing the bound intermediate, and [[3cts]] as the complex with the products. Positions of hydrogens on the ligands were calculated and added back to structures in the reaction scheme for instructional purposes and are not present in the experimentally-determined structures; additionally, arrows are drawn with atoms of the analog of acetyl-CoA to approximate the position of the reactive groups only as the reactive groups are not actually part of the analog or the molecules would have reacted; please, see the reaction scheme on this page for a more thorough accounting of the chemistry.</ref>. In this mechanism, three ionizable side chains in the active site of citrate synthase participate in acid-base catalysis: <scene name='Citrate_Synthase/5ctsactivesiteresidues/10'>His 274, His 320, and Asp 375</scene>. Citrate synthase is among one of the few enzymes that can directly form a carbon-carbon bond without the presence of metal ion cofactors.<br> | ||
*In step one, <scene name='Citrate_Synthase/5ctsasp375/9'>Asp 375 acts as a base removing a proton from the methyl group of acetyl-CoA</scene>, resulting in acetyl-CoA forming its enol; His 274 (magenta) stabilizes the acetyl-CoA enol by forming a hydrogen bond with the enol's oxygen. (See the reaction scheme below for a more thorough accounting of the chemistry.) | *In step one, <scene name='Citrate_Synthase/5ctsasp375/9'>Asp 375 acts as a base removing a proton from the methyl group of acetyl-CoA</scene>, resulting in acetyl-CoA forming its enol; His 274 (magenta) stabilizes the acetyl-CoA enol by forming a hydrogen bond with the enol's oxygen. (See the reaction scheme below for a more thorough accounting of the chemistry.) | ||
| Line 18: | Line 22: | ||
{{Button HeteroCPKPLUSCSColorScheme}} {{Link Toggle FancyCartoonHighQualityView}} | {{Button HeteroCPKPLUSCSColorScheme}} {{Link Toggle FancyCartoonHighQualityView}} | ||
| - | + | ==Regulation== | |
| + | |||
| + | Perhaps the most crucial regulators of the citrate synthase reaction are its substrates, acetyl-CoA and oxaloacetate. Both are present in the mitochondria at concentrations below saturation of citrate synthase. The metabolic flux is controlled by substrate availability, so controlling the levels of acetyl-CoA and oxaloacetate in the mitochondria controls the rate of reaction. Furthermore, citrate synthase is inhibited by NADH, <scene name='Citrate_Synthase/3ctscitrateonly/1'>citrate</scene> (which competes with oxaloacetate), and succinyl-CoA (an example of competitive feedback inhibition) <ref>PMID:3013232</ref>. In many plants, bacteria and fungi, such as the peroxisomes of baker's yeast, citrate synthase plays a role in the glyoxylate cycle<ref>PMID:3023912</ref><ref>PMID: 2181273</ref><ref>PMID:17615299</ref>. | ||
*<scene name='Citrate_Synthase/Cv/1'>Citrate Synthase Closed Form (Monomer)</scene> [[2cts]] | *<scene name='Citrate_Synthase/Cv/1'>Citrate Synthase Closed Form (Monomer)</scene> [[2cts]] | ||
*<scene name='Citrate_Synthase/Cv/2'>'Citrate Synthase Open Form (Monomer)</scene> [[1cts]] | *<scene name='Citrate_Synthase/Cv/2'>'Citrate Synthase Open Form (Monomer)</scene> [[1cts]] | ||
| Line 24: | Line 30: | ||
[[Image:CitSynReaction schematic as image.png|1024px|left|thumb| <span style="font-size:1.2em;">The reaction mechanism for catalysis by citrate synthase</span>]] | [[Image:CitSynReaction schematic as image.png|1024px|left|thumb| <span style="font-size:1.2em;">The reaction mechanism for catalysis by citrate synthase</span>]] | ||
==3D structures of Citrate Synthase== | ==3D structures of Citrate Synthase== | ||
| - | __NOTOC__ | ||
| Line 31: | Line 36: | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
[[Image:2cts plus overall reaction.png|500px|right|thumb| <span style="font-size:1.2em;">Citrate synthase 'closed' form ([[2cts]]) and the reaction</span>]] | [[Image:2cts plus overall reaction.png|500px|right|thumb| <span style="font-size:1.2em;">Citrate synthase 'closed' form ([[2cts]]) and the reaction</span>]] | ||
| - | + | {{#tree:id=OrganizedByTopic|openlevels=0| | |
| - | + | ||
| - | + | *Citrate synthase | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | **[[3msu]] – CitS – ''Francisella tularensis''<br /> | |
| + | **[[3enj]] – CitS – Wild boar<br /> | ||
| + | **[[2p2w]] – CitS – ''Thermotoga maritima''<br /> | ||
| + | **[[2c6x]] – CitS residues 2-364 – ''Bacillus subtilis''<br /> | ||
| + | **[[3l96]], [[1owc]], [[1nxe]], [[4jad]], [[4jae]] – EcCitS (mutant) – ''Eschericia coli''<br /> | ||
| + | **[[1k3p]], [[4g6b]] – EcCitS II<br /> | ||
| + | **[[2ibp]] – CitS – ''Pyrobaculum aerophilum''<br /> | ||
| + | **[[1iom]], [[1ixe]] – CitS – ''Thermus thermophilus''<br /> | ||
| + | **[[1o7x]] – CitS – ''Sulfolobus solfataricus''<br /> | ||
| + | **[[1a59]] – CitS – ''Antarctic bacterium''<br /> | ||
| + | **[[1aj8]] – CitS - ''Pyrococcus furiosus''<br /> | ||
| + | **[[5csc]], [[3cts]]– cCitS – chicken<br /> | ||
| + | **[[1cts]], [[2cts]] – pCitS - pig<br /> | ||
| + | **[[4e6y]] – CitS – ''Vibrio vulnificus'' | ||
| - | + | *Citrate synthase binary complex | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | **[[3l97]] - EcCitS (mutant) + S-carboxymethyl-CoA<br /> | |
| + | **[[2r9e]] – TaCitS + citryl dethia CoA – ''Thermoplasma acidophilum''<br /> | ||
| + | **[[6cts]] – cCitS + citryl thioether CoA<br /> | ||
| + | **[[3l98]], [[1owb]], [[1nxg]], [[4jaf]] - EcCitS (mutant) + NADH<br /> | ||
| + | **[[3l99]], [[4jag]] - EcCitS (mutant) + oxaloacetate<br /> | ||
| + | **[[2ifc]] - TaCitS + oxaloacetate | ||
| - | + | *Citrate synthase ternary complex | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | **[[2r26]] - TaCitS + oxaloacetate + S-carboxymethyl-CoA<br /> | ||
| + | **[[2h12]] - CitS + oxaloacetate + carboxymethyl dethia-CoA – ''Acetobacter aceti''<br /> | ||
| + | **[[1al6]], [[1csr]], [[1css]], [[1csh]], [[1csi]] - cCitS + oxaloacetate + carboxymethyl dethia-CoA derivative<br /> | ||
| + | **[[5cts]] - cCitS + oxaloacetate + carboxymethyl CoA<br /> | ||
| + | **[[1amz]] - cCitS + malate + nitromethyl dethia-CoA<br /> | ||
| + | **[[1csc]], [[2csc]], [[3csc]], [[4csc]] - cCitS + malate + carboxymethyl CoA<br /> | ||
| + | **[[6csc]] - cCitS + citrate + trifluoroacetonyl-CoA<br /> | ||
| + | **[[4cts]] - pCitS + oxaloacetate + S-acetonyl CoA<br /> | ||
| + | }} | ||
==See Also== | ==See Also== | ||
Revision as of 10:25, 20 November 2014
Contents |
3D structures of Citrate Synthase
Updated on 20-November-2014
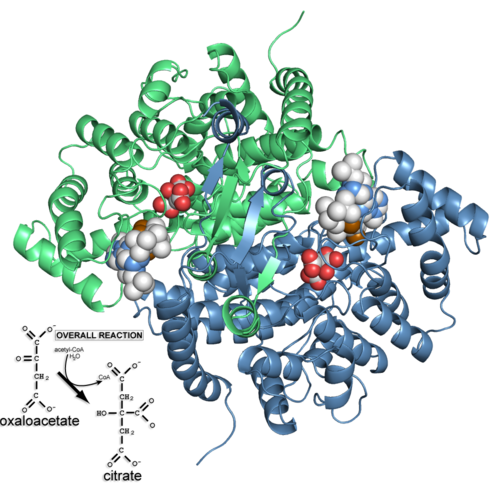
Citrate synthase 'closed' form (2cts) and the reaction
See Also
Literature and Notes
- ↑ "Citrate Synthase -." Wikipedia, the Free Encyclopedia. Web. 22 Mar. 2010.
- ↑ 2.0 2.1 Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008.
- ↑ 3.0 3.1 3.2 Remington S, Wiegand G, Huber R. Crystallographic refinement and atomic models of two different forms of citrate synthase at 2.7 and 1.7 A resolution. J Mol Biol. 1982 Jun 15;158(1):111-52. PMID:7120407
- ↑ In this structure 1cts, citrate, the resulting product of the conversion, is actually bound where oxaloacetate binds.
- ↑ Bayer E, Bauer B, Eggerer H. Evidence from inhibitor studies for conformational changes of citrate synthase. Eur J Biochem. 1981 Nov;120(1):155-60. PMID:7308213
- ↑ Karpusas M, Branchaud B, Remington SJ. Proposed mechanism for the condensation reaction of citrate synthase: 1.9-A structure of the ternary complex with oxaloacetate and carboxymethyl coenzyme A. Biochemistry. 1990 Mar 6;29(9):2213-9. PMID:2337600
- ↑ 5cts as the state preceding condensation with oxaloacetate and a non-reactive version of acetyl-CoA bound, 6cts as the state containing the bound intermediate, and 3cts as the complex with the products. Positions of hydrogens on the ligands were calculated and added back to structures in the reaction scheme for instructional purposes and are not present in the experimentally-determined structures; additionally, arrows are drawn with atoms of the analog of acetyl-CoA to approximate the position of the reactive groups only as the reactive groups are not actually part of the analog or the molecules would have reacted; please, see the reaction scheme on this page for a more thorough accounting of the chemistry.
- ↑ Wiegand G, Remington SJ. Citrate synthase: structure, control, and mechanism. Annu Rev Biophys Biophys Chem. 1986;15:97-117. PMID:3013232 doi:http://dx.doi.org/10.1146/annurev.bb.15.060186.000525
- ↑ Kim KS, Rosenkrantz MS, Guarente L. Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol Cell Biol. 1986 Jun;6(6):1936-42. PMID:3023912
- ↑ Lewin AS, Hines V, Small GM. Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol Cell Biol. 1990 Apr;10(4):1399-405. PMID:2181273
- ↑ Lee YJ, Hoe KL, Maeng PJ. Yeast cells lacking the CIT1-encoded mitochondrial citrate synthase are hypersusceptible to heat- or aging-induced apoptosis. Mol Biol Cell. 2007 Sep;18(9):3556-67. Epub 2007 Jul 5. PMID:17615299 doi:10.1091/mbc.E07-02-0118
External Resources
- Citrate Synthase: September 2007 Molecule of the Month as part of the series of tutorials that are at the RCSB Protein Data Bank and written by David Goodsell
- An interactive schematic animation of Citrate synthase's reaction mechanism from Lehninger's Principles of Biochemistry
- Citrate Synthase at Wikipedia
Proteopedia Page Contributors and Editors (what is this?)
Wayne Decatur, Michal Harel, Daniel Eddelman, Alexander Berchansky, Joel L. Sussman, Angel Herraez, David Canner, Eric Martz