We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Pheromone binding protein
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==Introduction== | ==Introduction== | ||
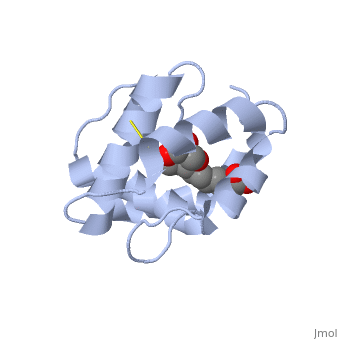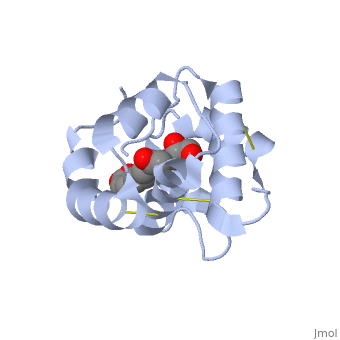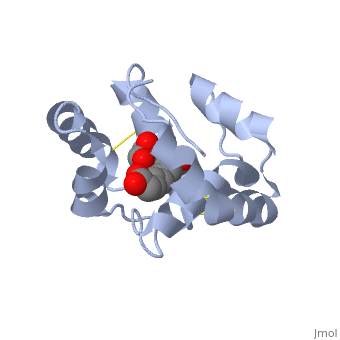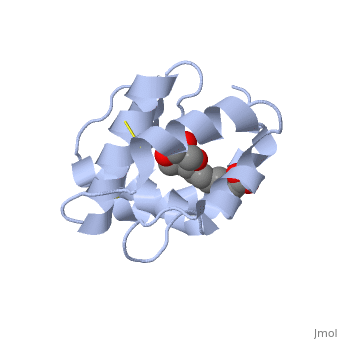
<StructureSection load='3bfa' size='340' side='right' caption='Pheromone binding protein of ''Apis mellifera'' scene=''> | <StructureSection load='3bfa' size='340' side='right' caption='Pheromone binding protein of ''Apis mellifera'' scene=''> | ||
| - | As the most ancient sense in nature, smell and chemical communication play a major role in successful mating, host and selection and other essential behaviors. | ||
| - | The detection of volatiles (often a small hydrophobic molecules) starts by | ||
| - | Pheromone binding proteins are soluble proteins | + | Pheromone binding proteins [http://en.wikipedia.org/wiki/Pheromone_binding_protein (PBP)] are type of Odorant binding proteins [http://en.wikipedia.org/wiki/Odorant-binding_protein (OBP)] - soluble proteins mediating the early stages of volatiles detection in both insects and vertebrates |
| - | As a model protein of this family we will further describe the well investigated Pheromone binding protein of the honey bee. | + | <ref>DOI:10.3389/fphys.2014.00320</ref>. |
| + | The volatiles (pheromones and other small hydrophobic molecules) are solubilized by the OBPs and activate the chemoreceptors. | ||
| + | |||
| + | As a model protein of this family we will further describe the well investigated Pheromone binding protein of the honey bee, ASP1. | ||
| + | |||
| + | == Pheromone-binding protein ASP1 == | ||
Chemical communication is crucial in social insects, where a complicated and delicate system of signals must be maintained in order to preserve the fragile equilibrium between the queen and the workers. | Chemical communication is crucial in social insects, where a complicated and delicate system of signals must be maintained in order to preserve the fragile equilibrium between the queen and the workers. | ||
| - | In the hive of the honey bee [http://en.wikipedia.org/wiki/Apis_mellifera ''Apis mellifera''] this equilibrium exists partially due to the extraction of blend of substances called queen mandibular pheromone [http://en.wikipedia.org/wiki/Honey_bee_pheromones#Queen_mandibular_pheromone (QMP)], by the queen. The three major component of the QMP blend are: 9-keto-2(E)-decenoic acid (9-ODA) and 9-hydroxy-2(E)-decenoic acid (9-HDA R-(−) or S-(+)). | + | In the hive of the honey bee [http://en.wikipedia.org/wiki/Apis_mellifera ''Apis mellifera''] this equilibrium exists partially due to the extraction of blend of substances called queen mandibular pheromone [http://en.wikipedia.org/wiki/Honey_bee_pheromones#Queen_mandibular_pheromone (QMP)], by the queen <ref>Winston, M.L., 1987. The Biology of the Honey Bee. Harvard University Press, Cambridge, MA.</ref>. The three major component of the QMP blend are: 9-keto-2(E)-decenoic acid (9-ODA) and 9-hydroxy-2(E)-decenoic acid (9-HDA R-(−) or S-(+)). |
| + | Pheromone-binding protein [http://www.uniprot.org/uniprot/Q9U9J6/ ASP] of the honeybee [http://en.wikipedia.org/wiki/Apis_mellifera ''Apis mellifera''] L. (Hymenoptera: Apidea) was first isolated and characterized by Danty ''et al''. (1998)<ref>DOI:10.1016/j.jmb.2008.04.048</ref> from the bee antennae. | ||
| + | Pheromone-binding protein [http://www.uniprot.org/uniprot/Q9U9J6/ ASP] of the honeybee [http://en.wikipedia.org/wiki/Apis_mellifera ''Apis mellifera''] L. (Hymenoptera: Apidea) was first isolated and characterized by Danty ''et al''. (1998)<ref>DOI:10.1016/j.jmb.2008.04.048</ref> from the bee antennae. | ||
| - | + | == Structure == | |
| - | <scene name='60/609542/Disulfide_bonds/1'>3 disulfide bonds</scene> | + | The protein is composed of 144 amino acids, which forms 6 alpha helices. Three <scene name='60/609542/Disulfide_bonds/1'>3 disulfide bonds</scene> tied four helices: disulfide 20–51 between H1 and H3, 47– 98 between H3 and H6, and 107–89 between H6 and H5. |
| + | |||
| + | == Interaction with the ligand 9-ODA== | ||
<scene name='60/609542/9-oda/3'>9-ODA</scene> | <scene name='60/609542/9-oda/3'>9-ODA</scene> | ||
| + | The carboxyl end of the component 9-ODA points towards the solvent, and has no any interaction with residues of the protein. The residues in the binding site are <scene name='60/609542/Binding_site/3'>hydrophobic</scene> | ||
| + | , and the connection between 9-ODA and ASP1 involve hydrogen bonds. | ||
| + | ---- | ||
| + | |||
<scene name='60/609542/Glycerol/2'>Glycerol</scene> | <scene name='60/609542/Glycerol/2'>Glycerol</scene> | ||
| - | <scene name='60/609542/Binding_site/3'>hydrophobic</scene> | ||
| - | Pheromone-binding protein [http://www.uniprot.org/uniprot/Q9U9J6/ ASP] of the honeybee [http://en.wikipedia.org/wiki/Apis_mellifera ''Apis mellifera''] L. (Hymenoptera: Apidea) was first isolated and characterized by Danty ''et al''. (1998)<ref>DOI:10.1016/j.jmb.2008.04.048</ref> from the bee antennae. | ||
or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
Revision as of 11:27, 22 November 2014
Introduction
| |||||||||||
References
- ↑ Pelosi P, Iovinella I, Felicioli A, Dani FR. Soluble proteins of chemical communication: an overview across arthropods. Front Physiol. 2014 Aug 27;5:320. doi: 10.3389/fphys.2014.00320. eCollection, 2014. PMID:25221516 doi:http://dx.doi.org/10.3389/fphys.2014.00320
- ↑ Winston, M.L., 1987. The Biology of the Honey Bee. Harvard University Press, Cambridge, MA.
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Tegoni M, Cambillau C. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J Mol Biol. 2008 Jun 27;380(1):158-69. Epub 2008 Apr 27. PMID:18508083 doi:10.1016/j.jmb.2008.04.048
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Tegoni M, Cambillau C. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J Mol Biol. 2008 Jun 27;380(1):158-69. Epub 2008 Apr 27. PMID:18508083 doi:10.1016/j.jmb.2008.04.048
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644