We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Molecular Playground/CLOCK:BMAL1 heterodimer complex
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Role in Circadian Rhythm == | == Role in Circadian Rhythm == | ||
| - | Circadian rhythms are operated by an endogenous core clock system that drives daily rhythms in behavior, physiology, and metabolism. In mammalian systems, the suprachiasmatic nucleus (SCN), which is located in the hypothalamus, is the locus of a master circadian clock. The SCN controls the expression of of proteins in a time dependent manner through a genetic feedback loop initiated by light passing through the eye.<ref>doi: 10.1111/ejn.12593</ref> The core molecular clockwork is composed of a transcriptional/post-translational feedback loop: CLOCK:BMAL1 (transcriptional activators) and PER:CRY (transcriptional repressors). In daytime, CLOCK and BMAL1 will form a heterodimer complex to bind the E-box promoter region of other circadian rhythm proteins, causing the transcription of Per (''Period'') and Cry (''Cryptochrome''). During the day, Per and Cry will dimerize and translocate into the nucleus, where they interact with CLOCK:BMAL1 to inhibit their own transcription, forming a negative feedback loop.<ref>DOI: 10.1126/science.1222804</ref> At night time, Per:Cry complex is degraded by a specific E3 ligase complex and the repression is relieved. After the repression level of Per:Cry is decreased, CLOCK:BMAL1 will be re-activated and start transcription again | + | Circadian rhythms are operated by an endogenous core clock system that drives daily rhythms in behavior, physiology, and metabolism. In mammalian systems, the suprachiasmatic nucleus (SCN), which is located in the hypothalamus, is the locus of a master circadian clock. The SCN controls the expression of of proteins in a time dependent manner through a genetic feedback loop initiated by light passing through the eye.<ref>doi: 10.1111/ejn.12593</ref> The core molecular clockwork is composed of a transcriptional/post-translational feedback loop: CLOCK:BMAL1 (transcriptional activators) and PER:CRY (transcriptional repressors). In daytime, CLOCK and BMAL1 will form a heterodimer complex to bind the E-box promoter region of other circadian rhythm proteins, causing the transcription of Per (''Period'') and Cry (''Cryptochrome''). During the day, Per and Cry will dimerize and translocate into the nucleus, where they interact with CLOCK:BMAL1 to inhibit their own transcription, forming a negative feedback loop.<ref>DOI: 10.1126/science.1222804</ref> At night time, Per:Cry complex is degraded by a specific E3 ligase complex and the repression is relieved. After the repression level of Per:Cry is decreased, CLOCK:BMAL1 will be re-activated and start transcription again, forming the positive feedback loop. The entire negative/positive feedback loop take around 24 hours to complete, thus forming the core mechanism of the circadian clock in mammals.<ref>doi:10.1016/B978-0-12-387690-4.00006-4</ref> |
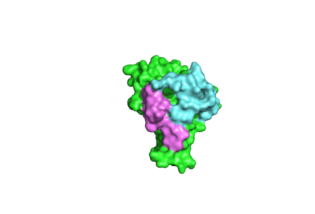
==The Overall structure of CLOCK:BMAL1 complex== | ==The Overall structure of CLOCK:BMAL1 complex== | ||
| - | The <scene name='60/609802/Clock_bmal1/1'>3D structure of CLOCK:BMAL1 heterodimer</scene> is shown in the right. It is a tightly | + | The <scene name='60/609802/Clock_bmal1/1'>3D structure of CLOCK:BMAL1 heterodimer</scene> is shown in the right. It is a tightly intertwined structure where CLOCK and BMAL1 are twisted together. Although the primary sequences of CLOCK and BMAL1 are similar, the structural arrangements of their domains are quite different. |
===CLOCK=== | ===CLOCK=== | ||
| - | <scene name='60/609802/Clock_only/1'>CLOCK</scene> is composed of three domains: one <scene name='60/609802/Clock_bhlh/1'>N-terminal bHLH</scene> domain, two PAS domains (<scene name='60/609802/Clock_psa_a/1'>PSA-A</scene> and <scene name='60/609802/Clock_psa_b/1'>PSA-B</scene>). The connections between each domain are two <scene name='60/609802/Clock_l1_l2/1'>flexible loops</scene>. | + | <scene name='60/609802/Clock_only/1'>CLOCK</scene> is composed of three domains: one <scene name='60/609802/Clock_bhlh/1'>N-terminal bHLH</scene> domain, two PAS domains (<scene name='60/609802/Clock_psa_a/1'>PSA-A</scene> and <scene name='60/609802/Clock_psa_b/1'>PSA-B</scene>). The connections between each domain are two <scene name='60/609802/Clock_l1_l2/1'>flexible loops</scene>. Comparedto the flexible loops in BMAL1, the distances of the connection loops in CLOCK are longer. |
===BMAL1=== | ===BMAL1=== | ||
| - | <scene name='60/609802/Bmal1_only/1'>BMAL1</scene> is also composed of three domains: one <scene name='60/609802/Bmal1_bhlh/1'>N-terminal bHLH</scene> domain, two PAS domains (<scene name='60/609802/Bmal1_psa_a/1'>PSA-A</scene> and <scene name='60/609802/Bmal1_psa_b/1'>PSA-B</scene>). There | + | <scene name='60/609802/Bmal1_only/1'>BMAL1</scene> is also composed of three domains: one <scene name='60/609802/Bmal1_bhlh/1'>N-terminal bHLH</scene> domain, two PAS domains (<scene name='60/609802/Bmal1_psa_a/1'>PSA-A</scene> and <scene name='60/609802/Bmal1_psa_b/1'>PSA-B</scene>). There is a ~15-residue flexible loop (<scene name='60/609802/Bmal1_l1/1'>L1</scene>) and a ~20-residue flexible loop (<scene name='60/609802/Bmal1_l2/1'>L2</scene>) connecting between each domain. |
==The interface between CLOCK and BMAL1== | ==The interface between CLOCK and BMAL1== | ||
| - | In the formation of CLOCK:BMAL1 heterodimer complex, each domain interacts with the corresponding domain of its partner subunit, | + | In the formation of the CLOCK:BMAL1 heterodimer complex, each domain interacts with the corresponding domain of its partner subunit, i.e. CLOCK bHLH interacts with BMAL1 bHLH domain and so forth. In PSA domains, <scene name='60/609802/Clock_bmal1_psa_a/1'>both CLOCK and BMAL1 PSA-A domains</scene> contain a five-stranded antiparallel β sheet and several α helices flanking the concave surface of the sheet. In those α helices, there are two <scene name='60/609802/Clock_bmal1_a_alpha/1'>N-terminal flanking helices (A'α) externals</scene> packed in between the β-sheet faces of the two domains to mediate the heterodimeric PSA-A interactions. |
=== PSA-A domain interface === | === PSA-A domain interface === | ||
| - | The interface | + | The interface bin the CLOCK:BMAL1 PSA-A dimer is mainly facilitated by hydrophobic interactions. Specifically, Phe104, Leu105, and Leu113 on <scene name='60/609802/Clock_bmal1_if_1/1'>the A'α helix of CLOCK contact the hydrophobic region on the β-sheet face of BMAL1</scene> (Leu159, Thr285, Tyr287, Val315, and Ile317). Similarly, Phe141, Leu142, and Leu150 on BMAL1 PSA-A contact the hydrophobic β-sheet face of CLOCK (F122, I216, V252, and T254). As a result, many residues obtained in the CLOCK:BMAL1 interface are conserved among bHLH-PAS transcription factors. This result may indicate that CLOCK and BMAL1 have a common PSA-A domain dimerization mode. |
===PSA-B domain interface=== | ===PSA-B domain interface=== | ||
| - | The PSA-B domains of CLOCK and BMAL1 are stacked in parallel conformation. The β-sheet on PSA-B domain of BMAL1 | + | The PSA-B domains of CLOCK and BMAL1 are stacked in parallel conformation. The β-sheet on PSA-B domain of BMAL1 contact the helical face of CLOCK PSA-B domain. The <scene name='60/609802/Clock_bmal1_if_2/1'>contact interface</scene> bury some hydrophobic residues on both subunits, including Try310, Val315, and Leu318 of CLOCK and Phe423, Trp427, and Val435 of BMAL1. |
===The binding interface between CLOCK:BMAL1 and E-box element=== | ===The binding interface between CLOCK:BMAL1 and E-box element=== | ||
Revision as of 00:38, 3 December 2014
CLOCK:BMAL1 heterodimer complex
| |||||||||||
References
- ↑ doi: https://dx.doi.org/10.1126/science.280.5369.1564
- ↑ Silver R, Kriegsfeld LJ. Circadian rhythms have broad implications for understanding brain and behavior. Eur J Neurosci. 2014 Jun;39(11):1866-80. doi: 10.1111/ejn.12593. Epub 2014 May 5. PMID:24799154 doi:http://dx.doi.org/10.1111/ejn.12593
- ↑ Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science. 2012 Jul 13;337(6091):189-94. Epub 2012 May 31. PMID:22653727 doi:10.1126/science.1222804
- ↑ Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175-230. doi: 10.1016/B978-0-12-387690-4.00006-4. PMID:21924978 doi:http://dx.doi.org/10.1016/B978-0-12-387690-4.00006-4
- ↑ Stevens RG. Circadian disruption and breast cancer: from melatonin to clock genes. Epidemiology. 2005 Mar;16(2):254-8. doi: 10.1097/01.ede.0000152525.21924.54. PMID:15703542 doi:http://dx.doi.org/10.1097/01.ede.0000152525.21924.54
- ↑ Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009 May 1;324(5927):651-4. doi: 10.1126/science.1171641. Epub 2009 Mar , 19. PMID:19299583 doi:http://dx.doi.org/10.1126/science.1171641
Proteopedia Page Contributors and Editors (what is this?)
Hui-Hsien Lin, Joseph Hardie, Michael Mingroni, Michal Harel