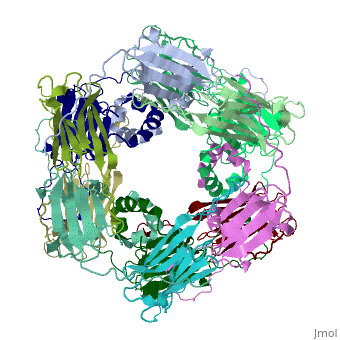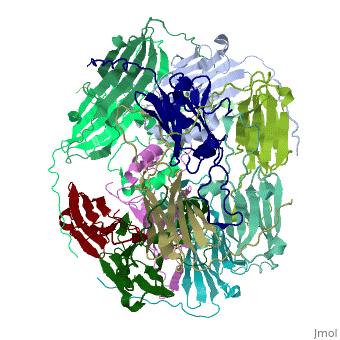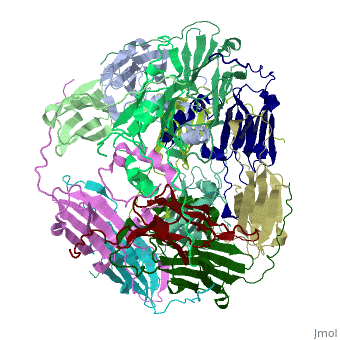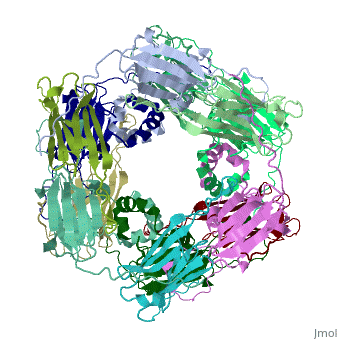Keithballard/sandbox
From Proteopedia
| Line 5: | Line 5: | ||
Molecular Playground banner: sHSP, a small but mighty protector against aggregation | Molecular Playground banner: sHSP, a small but mighty protector against aggregation | ||
| - | <scene name=' | + | <scene name='60/609774/Dimer_default/2'>Domain Architecture of sHSP Dimer</scene> |
| - | + | ||
| - | + | ||
| + | Small heat shock proteins (sHSPs) and related α-crystallins are virtually ubiquitous, ATP-independent molecular chaperones linked to diseases of protein misfolding. They comprise a conserved core α-crystallin domain (ACD - red) flanked by an evolutionarily variable N-terminal arm (NTA - green) and semi-conserved C-terminal extension (blue). They are capable of binding up to an equal mass of unfolding protein, forming large, heterogeneous sHSP-substrate complexes that make substrate available to the ATP-dependent chaperones for refolding. | ||
===Structure=== | ===Structure=== | ||
| + | The monomeric molecular weight of sHSPs range from 12-42 kDa. Many sHSPs form homo-oligomers consisting of 12 to >32 subunits per oligomer. The flexible NTA is disordered and is comprised of 3 small helices connected by random coils. The ACD core domain of the sHSP is an antiparellel β-sandwich containing the dimer interface, which is facilitated by a long loop participating in a strand exchange between partner monomers. The CTD contains an IXI motif (Ile147 and Ile 149), that patches the hydrophobic groove between the β4 and β8 strands in the interacting monomer. | ||
| - | + | ===Function=== | |
| - | + | sHSPs are believed to act as ATP independent molecular chaperones that are activated during proteotoxic stress by dissociating into an active form, presumably the sHSP dimer. This form has exposed hydrophobic regions which recognize and bind to hydrophobic patches on denaturing substrate protein. These interactions form a large, soluble, heterogeneous sHSP-substrate complex which can coordinate with ATP independent chaperones to refold substrate, or the degradation machinery for proteolysis. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 15:27, 3 December 2014
|
One of the CBI Molecules being studied in the University of Massachusetts Amherst Chemistry-Biology Interface Program at UMass Amherst and on display at the Molecular Playground.
Molecular Playground banner: sHSP, a small but mighty protector against aggregation
Small heat shock proteins (sHSPs) and related α-crystallins are virtually ubiquitous, ATP-independent molecular chaperones linked to diseases of protein misfolding. They comprise a conserved core α-crystallin domain (ACD - red) flanked by an evolutionarily variable N-terminal arm (NTA - green) and semi-conserved C-terminal extension (blue). They are capable of binding up to an equal mass of unfolding protein, forming large, heterogeneous sHSP-substrate complexes that make substrate available to the ATP-dependent chaperones for refolding.
Structure
The monomeric molecular weight of sHSPs range from 12-42 kDa. Many sHSPs form homo-oligomers consisting of 12 to >32 subunits per oligomer. The flexible NTA is disordered and is comprised of 3 small helices connected by random coils. The ACD core domain of the sHSP is an antiparellel β-sandwich containing the dimer interface, which is facilitated by a long loop participating in a strand exchange between partner monomers. The CTD contains an IXI motif (Ile147 and Ile 149), that patches the hydrophobic groove between the β4 and β8 strands in the interacting monomer.
Function
sHSPs are believed to act as ATP independent molecular chaperones that are activated during proteotoxic stress by dissociating into an active form, presumably the sHSP dimer. This form has exposed hydrophobic regions which recognize and bind to hydrophobic patches on denaturing substrate protein. These interactions form a large, soluble, heterogeneous sHSP-substrate complex which can coordinate with ATP independent chaperones to refold substrate, or the degradation machinery for proteolysis.