Pyrrolysyl-tRNA synthetase
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
<scene name='Sandbox_166/2ndstructure/3'>Showing the chains and bound BocLys</scene> ([[2zin]]). | <scene name='Sandbox_166/2ndstructure/3'>Showing the chains and bound BocLys</scene> ([[2zin]]). | ||
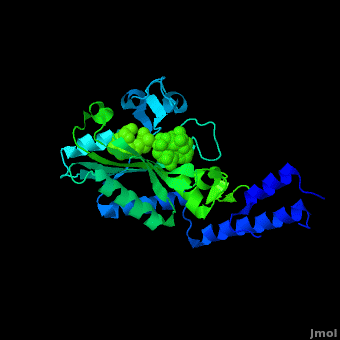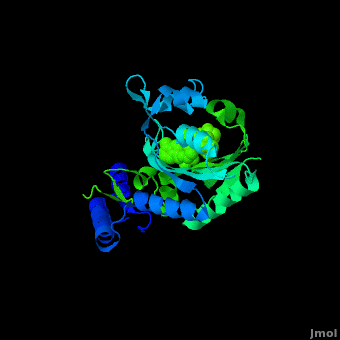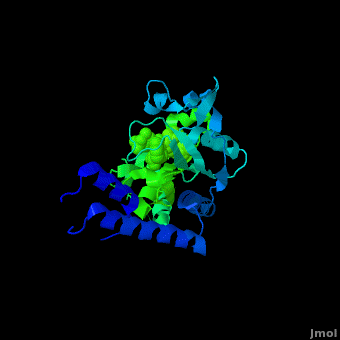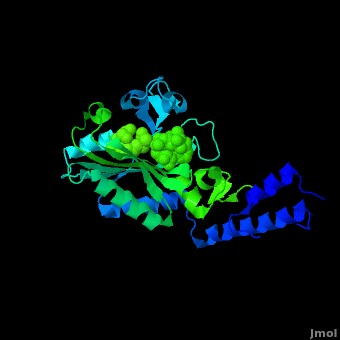
Pyrrolysyl-tRNA synthetase (PyIRS) catalytic complex attached with <scene name='Sandbox_166/2ndstructure/6'>BocLys</scene>, and along with adenosine 5’ (beta, gamma-imido) triphosphate (AMMPPNP) demonstrates the now known structural components and configuration that is needed for the efficient recognition of amino acids and the aminoacylation by PyIRS.<ref name="pept" /> As shown in figure 1, it consists of a multi domain polypeptide made of 1 chain (Chain A) comprising of a total length of 291 residues with 2 catalytic domains; PRK06253 and class_II_aaRS-like_core t.<ref name="pept" /> Furthermore, the structure is observed to consisting of 9 <scene name='Sandbox_166/2ndstructure/1'>α-helices</scene> (95 residues) making 32% of the structure, and the remaining is 12 <scene name='Sandbox_166/2ndstructure/2'>β-strands</scene> (59 residues) consisting of 20% of the other structure. In addition, its structure components require the presence of the 4 ligands; ANP, EDO, LBY, and MG for the protein to correctly perform its biological function. | Pyrrolysyl-tRNA synthetase (PyIRS) catalytic complex attached with <scene name='Sandbox_166/2ndstructure/6'>BocLys</scene>, and along with adenosine 5’ (beta, gamma-imido) triphosphate (AMMPPNP) demonstrates the now known structural components and configuration that is needed for the efficient recognition of amino acids and the aminoacylation by PyIRS.<ref name="pept" /> As shown in figure 1, it consists of a multi domain polypeptide made of 1 chain (Chain A) comprising of a total length of 291 residues with 2 catalytic domains; PRK06253 and class_II_aaRS-like_core t.<ref name="pept" /> Furthermore, the structure is observed to consisting of 9 <scene name='Sandbox_166/2ndstructure/1'>α-helices</scene> (95 residues) making 32% of the structure, and the remaining is 12 <scene name='Sandbox_166/2ndstructure/2'>β-strands</scene> (59 residues) consisting of 20% of the other structure. In addition, its structure components require the presence of the 4 ligands; ANP, EDO, LBY, and MG for the protein to correctly perform its biological function. | ||
| - | |||
PyIRS structure is found to enclose a hydrophobic interior where the catalytic activity of recognition and activation of the amino acids is going to take place.<ref name="paper"> PMID:18387634 </ref> This area creates a suitable environment for the binding to occur and for accommodating the residues of pyrrolysine methyl-pyrroline ring inside so it has the capability so further interact with the active-side residues available.<ref name="paper" /> It has been shown, that in order for the amino acid to successfully come in contact proper size of PyIRS must be incorporated at the N<sup>ϵ</sup>-carbonyl group.<ref name="pept" /> By using the Fo-Fc omit map and with the visible electron density in the active site it was experimentally trialed that the N<sup>ϵ</sup>-Boc group is situated in the hydrophobic interior in the similar was as the already observed traditional pyrrolysine AMPPNP bound arrangement.<ref name="pept" /> For that reason having the N<sup>ϵ</sup>-BocLys positioned in this way it has the capability to participate in the hydrogen bonding with Asn346 amide group.<ref name="pept" /> This Asn346 is important when the amino acid is binding because it helps attach its carbonyl side chain; as well as the main-chain α-amino group to ensure the proper recognition it going to take place.<ref name="paper" /> From this Asn346 will also allow the substrate to effectively bind to the side chain amide group by inducible fitting the carbonyl group of the substrate into position.<ref name="pept" /> Next, the Cα-carbonyl groups of BocLys in turn will hydrogen bond Asn346 contrary to the α-amino group which is linked to α-phosphate group of AMPPNP.<ref name="pept" /> Additionally, the BocLys α-carboxyl group is directed to that it is associated outside from the active site allowing for the flexibility due to the ability to rotate around the Cɑ- Cβ bond.<ref name="pept" /> Furthermore, this active site contains an abundant of functional residues such as Lys192, Arg197, Arg217, Lys336, Lys435, Lys438 and Arg439 from the one domain and Arg310, Lys311 and Arg314 from the other, which participate in the effective binding with the tRNA.<ref name="paper" /> | PyIRS structure is found to enclose a hydrophobic interior where the catalytic activity of recognition and activation of the amino acids is going to take place.<ref name="paper"> PMID:18387634 </ref> This area creates a suitable environment for the binding to occur and for accommodating the residues of pyrrolysine methyl-pyrroline ring inside so it has the capability so further interact with the active-side residues available.<ref name="paper" /> It has been shown, that in order for the amino acid to successfully come in contact proper size of PyIRS must be incorporated at the N<sup>ϵ</sup>-carbonyl group.<ref name="pept" /> By using the Fo-Fc omit map and with the visible electron density in the active site it was experimentally trialed that the N<sup>ϵ</sup>-Boc group is situated in the hydrophobic interior in the similar was as the already observed traditional pyrrolysine AMPPNP bound arrangement.<ref name="pept" /> For that reason having the N<sup>ϵ</sup>-BocLys positioned in this way it has the capability to participate in the hydrogen bonding with Asn346 amide group.<ref name="pept" /> This Asn346 is important when the amino acid is binding because it helps attach its carbonyl side chain; as well as the main-chain α-amino group to ensure the proper recognition it going to take place.<ref name="paper" /> From this Asn346 will also allow the substrate to effectively bind to the side chain amide group by inducible fitting the carbonyl group of the substrate into position.<ref name="pept" /> Next, the Cα-carbonyl groups of BocLys in turn will hydrogen bond Asn346 contrary to the α-amino group which is linked to α-phosphate group of AMPPNP.<ref name="pept" /> Additionally, the BocLys α-carboxyl group is directed to that it is associated outside from the active site allowing for the flexibility due to the ability to rotate around the Cɑ- Cβ bond.<ref name="pept" /> Furthermore, this active site contains an abundant of functional residues such as Lys192, Arg197, Arg217, Lys336, Lys435, Lys438 and Arg439 from the one domain and Arg310, Lys311 and Arg314 from the other, which participate in the effective binding with the tRNA.<ref name="paper" /> | ||
| Line 21: | Line 20: | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | + | *Pyrrolysyl-tRNA synthetase | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | **[[2e3c]] - MmPylRS catalytic domain – ''Methanosarcina mazei''<br /> | |
| + | **[[3vqw]], [[3vqx]] - MmPylRS catalytic domain (mutant)<br /> | ||
| + | **[[3dsq]] – DhPylRS - ''Desulfitobacterium hafniense'' | ||
| - | + | *Pyrrolysyl-tRNA synthetase binary complex | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | **[[3qtc]] – MmPylRS catalytic domain (mutant) + AMP-ATP analog<br /> | |
| + | **[[2q7e]], [[2zin]] - MmPylRS catalytic domain + ATP analog<br /> | ||
| + | **[[2zcd]] - MmPylRS catalytic domain + AMP-ATP analog<br /> | ||
| + | **[[3vqv]] - MmPylRS catalytic domain + AMPPNP <br /> | ||
| + | **[[4bw9]] - MmPylRS catalytic domain (mutant) + AMPPNP <br /> | ||
| + | **[[4bwa]] - MmPylRS catalytic domain (mutant) + adenylated norbornene <br /> | ||
| + | **[[2zni]] – DhPylRS + tRNA | ||
| - | + | *Pyrrolysyl-tRNA synthetase ternary complex | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | **[[2zce]] - MmPylRS catalytic domain + pyrrolysine + ATP analog<br /> | ||
| + | **[[2q7g]] - MmPylRS catalytic domain + pyrrolysine analog + ATP<br /> | ||
| + | **[[2zim]], [[2q7h]] - MmPylRS catalytic domain + adenylated pyrrolysine + pyrophosphate<br /> | ||
| + | **[[2zio]] - MmPylRS catalytic domain + AlocLys-AMP + ATP analog<br /> | ||
| + | **[[3vqy]] - MmPylRS catalytic domain + butoxycarbonyl lysine + AMPPNP <br /> | ||
| + | **[[2zin]] - MmPylRS catalytic domain + butoxycarbonyl lysine + ATP analog <br /> | ||
| + | }} | ||
==Additional Resources== | ==Additional Resources== | ||
For Additional information, see: [[Translation]]<br /> | For Additional information, see: [[Translation]]<br /> | ||
Revision as of 09:17, 7 December 2014
| |||||||||||
3D structures of pyrrolysyl-tRNA synthetase
Updated on 07-December-2014
Additional Resources
For Additional information, see: Translation
References
- ↑ 1.0 1.1 Herring S, Ambrogelly A, Polycarpo CR, Soll D. Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 2007;35(4):1270-8. Epub 2007 Jan 31. PMID:17267409 doi:10.1093/nar/gkl1151
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem Biol. 2008 Nov 24;15(11):1187-97. PMID:19022179 doi:10.1016/j.chembiol.2008.10.004
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Urbancsek J, Rabe T, Grunwald K, Kiesel L, Papp Z, Runnebaum B. High preovulatory serum luteinizing hormone level is unfavorable to conception. Gynecol Endocrinol. 1991 Dec;5(4):223-33. PMID:1796745
- ↑ Polycarpo C, Ambrogelly A, Berube A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Soll D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12450-4. Epub 2004 Aug 16. PMID:15314242 doi:10.1073/pnas.0405362101
- ↑ 5.0 5.1 5.2 Nozawa K, O'Donoghue P, Gundllapalli S, Araiso Y, Ishitani R, Umehara T, Soll D, Nureki O. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature. 2009 Feb 26;457(7233):1163-7. Epub 2008 Dec 31. PMID:19118381 doi:10.1038/nature07611
- ↑ 6.0 6.1 Soares JA, Zhang L, Pitsch RL, Kleinholz NM, Jones RB, Wolff JJ, Amster J, Green-Church KB, Krzycki JA. The residue mass of L-pyrrolysine in three distinct methylamine methyltransferases. J Biol Chem. 2005 Nov 4;280(44):36962-9. Epub 2005 Aug 11. PMID:16096277 doi:10.1074/jbc.M506402200
- ↑ Polycarpo CR, Herring S, Berube A, Wood JL, Soll D, Ambrogelly A. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006 Dec 11;580(28-29):6695-700. Epub 2006 Nov 20. PMID:17126325 doi:10.1016/j.febslet.2006.11.028
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J Mol Biol. 2008 May 2;378(3):634-52. Epub 2008 Feb 29. PMID:18387634 doi:10.1016/j.jmb.2008.02.045
- ↑ 9.0 9.1 9.2 9.3 Ibba M, Soll D. Genetic code: introducing pyrrolysine. Curr Biol. 2002 Jul 9;12(13):R464-6. PMID:12121639
Proteopedia Page Contributors and Editors (what is this?)
Agatka Jaworski, Michal Harel, Joel L. Sussman, David Canner, Andrea Gorrell, Alexander Berchansky