Trypsin
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
__TOC__ | __TOC__ | ||
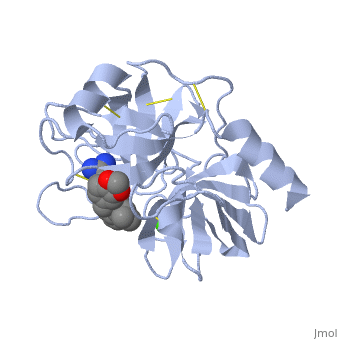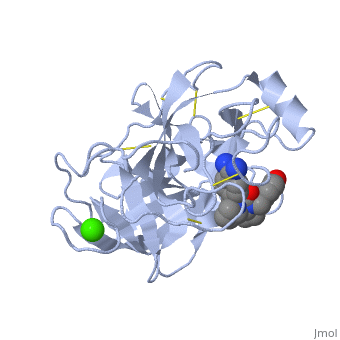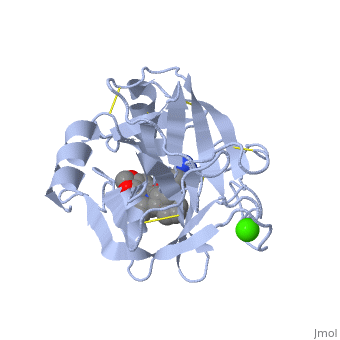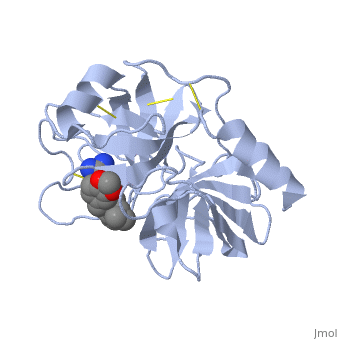
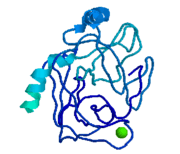
| - | '''Trypsin''' is a medium size globular protein that functions as a pancreatic serine protease. This enzyme hydrolyzes bonds by cleaving peptides on the C-terminal side of the amino acid residues lysine and arginine. It has also been shown that cleavage will not occur if there is a proline residue on the carboxyl side of the cleavage site. Trypsin was first discovered in 1876 by Kuhne, who investigated the proteolytic activity of the enzyme. In 1931 the enzyme was purified by crystallization by Norothrop and Kunitz and later in 1974 the three dimensional structure of trypsin was determined. Throughout the 1990's the role of trypsin in hereditary pancreatitis and the mutation that causes it was discovered. Today trypsin is used in the development of cell and tissue protocols, as well as in the medical field to determine the role of trypsin in pancreatic diseases<ref>Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html></ref>. | + | *'''Trypsin''' is a medium size globular protein that functions as a pancreatic serine protease. This enzyme hydrolyzes bonds by cleaving peptides on the C-terminal side of the amino acid residues lysine and arginine. It has also been shown that cleavage will not occur if there is a proline residue on the carboxyl side of the cleavage site. Trypsin was first discovered in 1876 by Kuhne, who investigated the proteolytic activity of the enzyme. In 1931 the enzyme was purified by crystallization by Norothrop and Kunitz and later in 1974 the three dimensional structure of trypsin was determined. Throughout the 1990's the role of trypsin in hereditary pancreatitis and the mutation that causes it was discovered. Today trypsin is used in the development of cell and tissue protocols, as well as in the medical field to determine the role of trypsin in pancreatic diseases<ref>Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html></ref>. |
[[Image:Tryogen.gif |thumb|left|Trypsinogen]] | [[Image:Tryogen.gif |thumb|left|Trypsinogen]] | ||
{{Clear}} | {{Clear}} | ||
| - | Trypsinogen is the precursor form or zymogen of Trypsin. Zymogens are enzyme precursors that are made and secreted in the lysosome of the cell. Zymogens are not active until they go through a chemical process such as hydrolysis, cleavage, or other cleavages that reveal the active site. The zymogen precursor is necessary in order to prevent the destruction of cellular proteins and to allow the enzyme to be in it's active state only when in appropriate conditions. Trypsinogen is specifically produced in the exocrine cells of the pancreas. There are three isoforms of Trypsinogen that are secreted by the pancreas. The precursor is only activated when it reaches the lumen of the small intestine. This activation occurs through the help of an enteropeptidase and once activated trypsin stimulates the formation of more trypsinogen<ref>Trypsin. 30 October 2010 <http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzyme/trypsin.html>.</ref>. The structure of Bovine Trypsinogen is shown in the figure to the right <ref> Image From: http://chemistry.umeche.maine.edu/MAT500/Peptidase1.html</ref>. | + | *'''Trypsinogen''' is the precursor form or zymogen of Trypsin. Zymogens are enzyme precursors that are made and secreted in the lysosome of the cell. Zymogens are not active until they go through a chemical process such as hydrolysis, cleavage, or other cleavages that reveal the active site. The zymogen precursor is necessary in order to prevent the destruction of cellular proteins and to allow the enzyme to be in it's active state only when in appropriate conditions. Trypsinogen is specifically produced in the exocrine cells of the pancreas. There are three isoforms of Trypsinogen that are secreted by the pancreas. The precursor is only activated when it reaches the lumen of the small intestine. This activation occurs through the help of an enteropeptidase and once activated trypsin stimulates the formation of more trypsinogen<ref>Trypsin. 30 October 2010 <http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzyme/trypsin.html>.</ref>. The structure of Bovine Trypsinogen is shown in the figure to the right <ref> Image From: http://chemistry.umeche.maine.edu/MAT500/Peptidase1.html</ref>. |
Trypsin has many applications due to fact that it is easily purified in high quantities. The trypsin enzyme is often used in the research setting to digest proteins and then identify the resulting peptides using mass spectrometry. Trypsin has many uses in the medical field such as dissolving blood clots and treating inflammation. Other applications include its use in pre-digesting of baby food, fingerprinting and sequencing work, and environmental monitoring <ref> Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html></ref>. For additional details see [[Serine Proteases]]. | Trypsin has many applications due to fact that it is easily purified in high quantities. The trypsin enzyme is often used in the research setting to digest proteins and then identify the resulting peptides using mass spectrometry. Trypsin has many uses in the medical field such as dissolving blood clots and treating inflammation. Other applications include its use in pre-digesting of baby food, fingerprinting and sequencing work, and environmental monitoring <ref> Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html></ref>. For additional details see [[Serine Proteases]]. | ||
| Line 40: | Line 40: | ||
Trypsin, chymotrypsin, and elastase are all digestive enzymes that are produced in the pancreas and catalyze the hydrolysis of peptide bonds. Each of these enzymes has different specificities in regards to the side chains next to the peptide bond. Chymotrypsin prefers a large hydrophobic residue, trypsin is specific for a positively charged residue, and elastase prefers a small neutral residue. Chymotrypsin, trypsin and elastase are all proteins that contain a catalytic mechanism and hydrolyze peptides using the serine protease mechanism. Chymotrypsin and elastase are both homologs of Trypsin since they are 40% alike in structure and composition <ref> Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008. </ref>. In the <scene name='Sandbox_32/Chymotrypsin/2'>Chymotrypsin</scene> structure shown the alpha helices are blue, the beta sheets are green, and the remainder of the protein is red. In the <scene name='Sandbox_32/Elastase/2'>Elastase</scene> structure shown the alpha helices are in red, the beta sheets are yellow, and the remainder of the protein is orange. | Trypsin, chymotrypsin, and elastase are all digestive enzymes that are produced in the pancreas and catalyze the hydrolysis of peptide bonds. Each of these enzymes has different specificities in regards to the side chains next to the peptide bond. Chymotrypsin prefers a large hydrophobic residue, trypsin is specific for a positively charged residue, and elastase prefers a small neutral residue. Chymotrypsin, trypsin and elastase are all proteins that contain a catalytic mechanism and hydrolyze peptides using the serine protease mechanism. Chymotrypsin and elastase are both homologs of Trypsin since they are 40% alike in structure and composition <ref> Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008. </ref>. In the <scene name='Sandbox_32/Chymotrypsin/2'>Chymotrypsin</scene> structure shown the alpha helices are blue, the beta sheets are green, and the remainder of the protein is red. In the <scene name='Sandbox_32/Elastase/2'>Elastase</scene> structure shown the alpha helices are in red, the beta sheets are yellow, and the remainder of the protein is orange. | ||
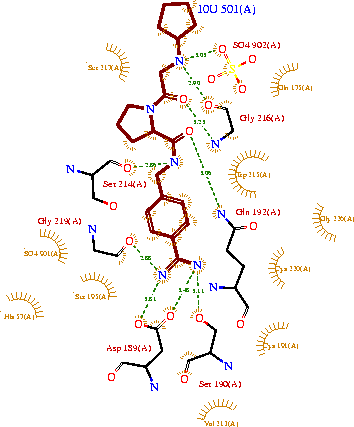
| - | + | '''The remarkable efficiency of a Pin-II proteinase inhibitor sans two conserved disulfide bonds is due to enhanced flexibility and hydrogen-bond density in the reactive loop''' <ref>doi 10.1080/07391102.2012.745378</ref> | |
Background: Plant proteinase Inhibitors (PIs) are ubiquitous in the plant kingdom and have been extensively studied as plant defense molecules, which inhibit hydrolytic enzymes (''e.g.'' <scene name='Journal:JBSD:39/Cv/12'>trypsin</scene>, <font color='darkmagenta'><b>colored in darkmagenta</b></font>) of the insect gut <ref name="Green">PMID: 17836138</ref>. Among various PI families, Serine PI Pin-II/Pot-II family displays a remarkable structural and functional diversity at the gene and protein level <ref name="Kong">PMID: 18315854</ref>. Wound, herbivory and stress induced up-regulation of these PIs clearly link them to plant defense <ref name="Green">PMID: 17836138</ref>. Previous studies using transgenic systems or in vivo assays have positively correlated the advantage offered by Pin-II PI expression in plants against insect attack <ref name="Johnson">PMID: 2602379</ref> <ref name="Duan">PMID: 9630927</ref>. Precursor proteins of Pin-II PIs consist of 1- to 8- <scene name='Journal:JBSD:39/Cv/4'>inhibitory repeat domains (IRDs)</scene> connected by proteolytic-sensitive linkers, which releases IRD units upon cleavage. <scene name='Journal:JBSD:39/Cv/5'>Each IRD is a peptide of around 50 aa length</scene> (<span style="color:lime;background-color:black;font-weight:bold;">colored in green</span>) with a molecular mass of ~6 KDa. The aa sequence of IRDs shows variations, at the same time the <scene name='Journal:JBSD:39/Cv/6'>8 cysteine residues that form disulfide bridge are conserved</scene> (<span style="color:yellow;background-color:black;font-weight:bold;">colored in yellow</span>) <ref name="Nielsen">PMID: 7578034</ref> <ref name="Scanlon">PMID: 10425681</ref> <ref name="Lee">PMID: 10360353</ref> <ref name="Schirra">PMID: 11178894</ref>. One structural feature of Pin-II IRD is a disordered loop with triple stranded β sheet scaffold. The disordered solvent exposed reactive loop is anchored by the four conserved disulfide bonds (C4-C41, C7-C25, C8-C37 and C14-C50) <ref name="Schirra1">PMID: 16029154</ref> <ref name="Schirra2">PMID: 18991765</ref>. Among the four disulfide bonds, C8-C37 has been found to be very crucial for maintaining active conformation, whereas C4-C41 has an important role in maintaining the flexibility of the reactive loop <ref name="Schirra3">PMID: 19925809</ref>. Thus, any selective loss of disulfide bond is expected to have evolutionary significance leading to functional differentiation of inhibitors <ref name="Li">PMID: 21494600</ref>. | Background: Plant proteinase Inhibitors (PIs) are ubiquitous in the plant kingdom and have been extensively studied as plant defense molecules, which inhibit hydrolytic enzymes (''e.g.'' <scene name='Journal:JBSD:39/Cv/12'>trypsin</scene>, <font color='darkmagenta'><b>colored in darkmagenta</b></font>) of the insect gut <ref name="Green">PMID: 17836138</ref>. Among various PI families, Serine PI Pin-II/Pot-II family displays a remarkable structural and functional diversity at the gene and protein level <ref name="Kong">PMID: 18315854</ref>. Wound, herbivory and stress induced up-regulation of these PIs clearly link them to plant defense <ref name="Green">PMID: 17836138</ref>. Previous studies using transgenic systems or in vivo assays have positively correlated the advantage offered by Pin-II PI expression in plants against insect attack <ref name="Johnson">PMID: 2602379</ref> <ref name="Duan">PMID: 9630927</ref>. Precursor proteins of Pin-II PIs consist of 1- to 8- <scene name='Journal:JBSD:39/Cv/4'>inhibitory repeat domains (IRDs)</scene> connected by proteolytic-sensitive linkers, which releases IRD units upon cleavage. <scene name='Journal:JBSD:39/Cv/5'>Each IRD is a peptide of around 50 aa length</scene> (<span style="color:lime;background-color:black;font-weight:bold;">colored in green</span>) with a molecular mass of ~6 KDa. The aa sequence of IRDs shows variations, at the same time the <scene name='Journal:JBSD:39/Cv/6'>8 cysteine residues that form disulfide bridge are conserved</scene> (<span style="color:yellow;background-color:black;font-weight:bold;">colored in yellow</span>) <ref name="Nielsen">PMID: 7578034</ref> <ref name="Scanlon">PMID: 10425681</ref> <ref name="Lee">PMID: 10360353</ref> <ref name="Schirra">PMID: 11178894</ref>. One structural feature of Pin-II IRD is a disordered loop with triple stranded β sheet scaffold. The disordered solvent exposed reactive loop is anchored by the four conserved disulfide bonds (C4-C41, C7-C25, C8-C37 and C14-C50) <ref name="Schirra1">PMID: 16029154</ref> <ref name="Schirra2">PMID: 18991765</ref>. Among the four disulfide bonds, C8-C37 has been found to be very crucial for maintaining active conformation, whereas C4-C41 has an important role in maintaining the flexibility of the reactive loop <ref name="Schirra3">PMID: 19925809</ref>. Thus, any selective loss of disulfide bond is expected to have evolutionary significance leading to functional differentiation of inhibitors <ref name="Li">PMID: 21494600</ref>. | ||
Revision as of 12:17, 15 December 2014
| |||||||||||
3D structures of Trypsin
Updated on 15-December-2014
References
- ↑ Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html>
- ↑ Trypsin. 30 October 2010 <http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzyme/trypsin.html>.
- ↑ Image From: http://chemistry.umeche.maine.edu/MAT500/Peptidase1.html
- ↑ Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html>
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
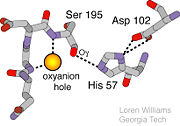
- ↑ Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.
- ↑ Image From:

- ↑ Williams, Loren. Georgia Tech. http://www2.chemistry.gatech.edu/~1W26/bcourse_information/6521/protein/serine_protease/triad_1/html.
- ↑ Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
- ↑ Joshi RS, Mishra M, Tamhane VA, Ghosh A, Sonavane U, Suresh CG, Joshi R, Gupta VS, Giri AP. The remarkable efficiency of a Pin-II proteinase inhibitor sans two conserved disulfide bonds is due to enhanced flexibility and hydrogen bond density in the reactive site loop. J Biomol Struct Dyn. 2012 Dec 20. PMID:23256852 doi:10.1080/07391102.2012.745378
- ↑ 12.0 12.1 Green TR, Ryan CA. Wound-Induced Proteinase Inhibitor in Plant Leaves: A Possible Defense Mechanism against Insects. Science. 1972 Feb 18;175(4023):776-7. PMID:17836138 doi:10.1126/science.175.4023.776
- ↑ Kong L, Ranganathan S. Tandem duplication, circular permutation, molecular adaptation: how Solanaceae resist pests via inhibitors. BMC Bioinformatics. 2008;9 Suppl 1:S22. PMID:18315854 doi:10.1186/1471-2105-9-S1-S22
- ↑ Johnson R, Narvaez J, An G, Ryan C. Expression of proteinase inhibitors I and II in transgenic tobacco plants: effects on natural defense against Manduca sexta larvae. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9871-5. PMID:2602379
- ↑ Duan X, Li X, Xue Q, Abo-el-Saad M, Xu D, Wu R. Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat Biotechnol. 1996 Apr;14(4):494-8. PMID:9630927 doi:10.1038/nbt0496-494
- ↑ Nielsen KJ, Heath RL, Anderson MA, Craik DJ. Structures of a series of 6-kDa trypsin inhibitors isolated from the stigma of Nicotiana alata. Biochemistry. 1995 Nov 7;34(44):14304-11. PMID:7578034
- ↑ Scanlon MJ, Lee MC, Anderson MA, Craik DJ. Structure of a putative ancestral protein encoded by a single sequence repeat from a multidomain proteinase inhibitor gene from Nicotiana alata. Structure. 1999 Jul 15;7(7):793-802. PMID:10425681
- ↑ Lee MC, Scanlon MJ, Craik DJ, Anderson MA. A novel two-chain proteinase inhibitor generated by circularization of a multidomain precursor protein. Nat Struct Biol. 1999 Jun;6(6):526-30. PMID:10360353 doi:10.1038/9293
- ↑ Schirra HJ, Scanlon MJ, Lee MC, Anderson MA, Craik DJ. The solution structure of C1-T1, a two-domain proteinase inhibitor derived from a circular precursor protein from Nicotiana alata. J Mol Biol. 2001 Feb 9;306(1):69-79. PMID:11178894 doi:10.1006/jmbi.2000.4318
- ↑ Schirra HJ, Craik DJ. Structure and folding of potato type II proteinase inhibitors: circular permutation and intramolecular domain swapping. Protein Pept Lett. 2005 Jul;12(5):421-31. PMID:16029154
- ↑ Schirra HJ, Anderson MA, Craik DJ. Structural refinement of insecticidal plant proteinase inhibitors from Nicotiana alata. Protein Pept Lett. 2008;15(9):903-9. PMID:18991765
- ↑ Schirra HJ, Guarino RF, Anderson MA, Craik DJ. Selective removal of individual disulfide bonds within a potato type II serine proteinase inhibitor from Nicotiana alata reveals differential stabilization of the reactive-site loop. J Mol Biol. 2010 Jan 22;395(3):609-26. Epub 2009 Nov 17. PMID:19925809 doi:10.1016/j.jmb.2009.11.031
- ↑ Li XQ, Zhang T, Donnelly D. Selective loss of cysteine residues and disulphide bonds in a potato proteinase inhibitor II family. PLoS One. 2011 Apr 11;6(4):e18615. PMID:21494600 doi:10.1371/journal.pone.0018615
- ↑ Barrette-Ng IH, Ng KK, Cherney MM, Pearce G, Ryan CA, James MN. Structural basis of inhibition revealed by a 1:2 complex of the two-headed tomato inhibitor-II and subtilisin Carlsberg. J Biol Chem. 2003 Jun 27;278(26):24062-71. Epub 2003 Apr 8. PMID:12684499 doi:10.1074/jbc.M302020200
- ↑ Dunse KM, Kaas Q, Guarino RF, Barton PA, Craik DJ, Anderson MA. Molecular basis for the resistance of an insect chymotrypsin to a potato type II proteinase inhibitor. Proc Natl Acad Sci U S A. 2010 Aug 24;107(34):15016-21. Epub 2010 Aug 9. PMID:20696921 doi:10.1073/pnas.1009327107
- ↑ Tamhane VA, Giri AP, Kumar P, Gupta VS. Spatial and temporal expression patterns of diverse Pin-II proteinase inhibitor genes in Capsicum annuum Linn. Gene. 2009 Aug 1;442(1-2):88-98. Epub 2009 Apr 22. PMID:19393726 doi:10.1016/j.gene.2009.04.012
- ↑ Tamhane VA, Chougule NP, Giri AP, Dixit AR, Sainani MN, Gupta VS. In vivo and in vitro effect of Capsicum annum proteinase inhibitors on Helicoverpa armigera gut proteinases. Biochim Biophys Acta. 2005 Mar 11;1722(2):156-67. Epub 2005 Jan 12. PMID:15715970 doi:10.1016/j.bbagen.2004.12.017
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Eran Hodis, Leah Bowlin, David Canner, Karsten Theis, Glenn Jones, Ben Hallowell, Karl Oberholser, Jaime Prilusky