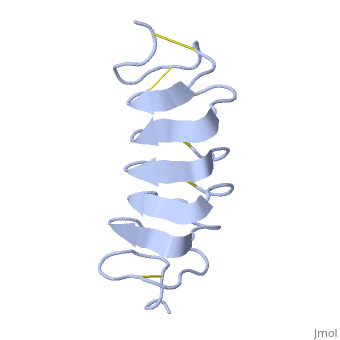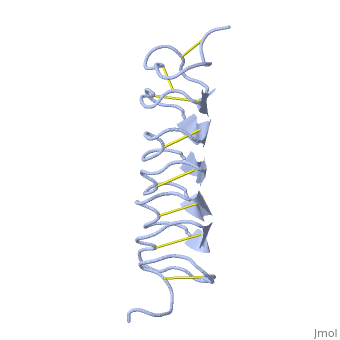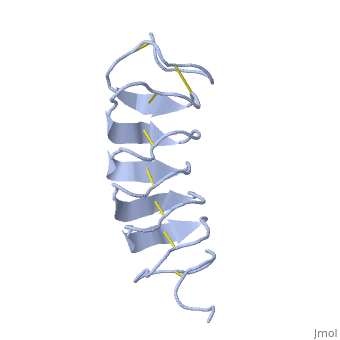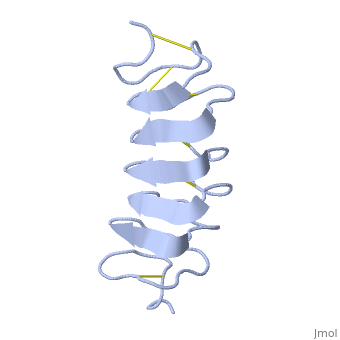
Tenebrio molitor Antifreeze Protein (TmAFP)
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
TmAFP is the smallest Beta-Helix with only 12 amino acids per turn. Therefore, it has a very narrow bore, which is constricted and further bisected by disulphide bonds to form two channels, leaving no room for hydrophobic core. | TmAFP is the smallest Beta-Helix with only 12 amino acids per turn. Therefore, it has a very narrow bore, which is constricted and further bisected by disulphide bonds to form two channels, leaving no room for hydrophobic core. | ||
| - | The few Hydrophobic residues <scene name='61/612804/Hydrophobic/1'>val25, val34, phe58, tyr70</scene> have their side chains projecting outwards to the solvent. In the core there is room only for the relatively small side chains of the conserves Serine and Alanine to project into the core, on either side of the bisecting disulphide bridge. | + | The few Hydrophobic residues <scene name='61/612804/Hydrophobic/1'>val25, val34, phe58, tyr70</scene> have their side chains projecting outwards to the solvent. In the core there is room only for the relatively small side chains of the conserves Serine and Alanine to project into the core, on either side of the bisecting disulphide bridge.<ref>DOI 10.1038/35018604</ref> |
| Line 20: | Line 20: | ||
| - | + | or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | |
== Function == | == Function == | ||
| Line 39: | Line 39: | ||
The regular array formed by this 3 ranks of oxygen atoms can be seen as a small piece of one layer thick ice to be incorporated into a large ice lattice. | The regular array formed by this 3 ranks of oxygen atoms can be seen as a small piece of one layer thick ice to be incorporated into a large ice lattice. | ||
| - | There is no need to readjust Threonine side chains, because they don’t present steric interference. | + | There is no need to readjust Threonine side chains, because they don’t present steric interference. <ref>DOI 10.1038/35018604</ref> |
Revision as of 14:44, 16 December 2014
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Liou YC, Tocilj A, Davies PL, Jia Z. Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein. Nature. 2000 Jul 20;406(6793):322-4. PMID:10917536 doi:10.1038/35018604
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Liou YC, Tocilj A, Davies PL, Jia Z. Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein. Nature. 2000 Jul 20;406(6793):322-4. PMID:10917536 doi:10.1038/35018604