Vascular Endothelial Growth Factor Receptor
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
{{Clear}} | {{Clear}} | ||
| + | ==Introduction== | ||
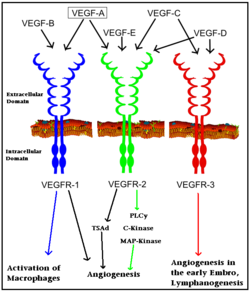
[[Vascular Endothelial Growth Factor Receptor]]s (VEGFRs) are tyrosine kinase receptors responsible for binding with [[VEGF]] to initiate signal cascades that stimulate angiogenesis among other effects. VEGFRs convey signals to other signal transduction effectors via autophosphorylation of specific residues in its structure. Because VEGFRs are up-regulated in cancerous tumors which have a high metabolic need for oxygen, VEGFRs are an important target for [[pharmaceutical drugs]] treating [[cancer]]. VEGFR subtypes are numbered 1,2,3. | [[Vascular Endothelial Growth Factor Receptor]]s (VEGFRs) are tyrosine kinase receptors responsible for binding with [[VEGF]] to initiate signal cascades that stimulate angiogenesis among other effects. VEGFRs convey signals to other signal transduction effectors via autophosphorylation of specific residues in its structure. Because VEGFRs are up-regulated in cancerous tumors which have a high metabolic need for oxygen, VEGFRs are an important target for [[pharmaceutical drugs]] treating [[cancer]]. VEGFR subtypes are numbered 1,2,3. | ||
[[Image: VEGF_receptors.png|250px|left|thumb| Interaction of VEGFs with VEGFRs. Colored arrows indicate major pathway. Black arrows indicate minor pathway.]] | [[Image: VEGF_receptors.png|250px|left|thumb| Interaction of VEGFs with VEGFRs. Colored arrows indicate major pathway. Black arrows indicate minor pathway.]] | ||
{{Clear}} | {{Clear}} | ||
| - | ==Biological Function | + | ==Biological Function == |
The VEGFRs are a family of tyrosine kinase receptors on the surface of different cells depending on family identity. VEGFR-1 is expressed on haematopoietic stem cells, monocytes, and vascular endothelial cells. VEGFR-2 is expressed on vascular endothelial cells and lymphatic endothelial cells, while VEGFR-3 is only expressed on lymphatic endothelial cells.<ref>PMID:16633338</ref> | The VEGFRs are a family of tyrosine kinase receptors on the surface of different cells depending on family identity. VEGFR-1 is expressed on haematopoietic stem cells, monocytes, and vascular endothelial cells. VEGFR-2 is expressed on vascular endothelial cells and lymphatic endothelial cells, while VEGFR-3 is only expressed on lymphatic endothelial cells.<ref>PMID:16633338</ref> | ||
| Line 12: | Line 13: | ||
In terms of function, VEGFR-1 is required for the recruitment of haematopoietic stem cells as well as the migration of monocytes and macrophages while VEGFR-2 regulates vascular endothelial function and VEGFR-3 regulates lymphatic endothelial cell function.<ref>PMID: 17658244</ref> VEGFR-2 has been the focus of the most research as it is the major signal transducer of both physioligcal, and perhaps more importantly, pathological angiogenesis, especially in cancerous tumors. VEGFR-2 is of critical importance to the body as exemplified by Shalaby ''et al.'' who demonstrated that VEGFR-2 gene knockout mice die at E8-8.5 due to lack of vasculogenesis.<ref>PMID:7596453</ref> The signal cascade initiated by binding VEGF to VEGFR is dependent upon specific sites of phosphorylation in the VEGFR structure and the interaction between these phosphorylated sites and other signaling molecules. | In terms of function, VEGFR-1 is required for the recruitment of haematopoietic stem cells as well as the migration of monocytes and macrophages while VEGFR-2 regulates vascular endothelial function and VEGFR-3 regulates lymphatic endothelial cell function.<ref>PMID: 17658244</ref> VEGFR-2 has been the focus of the most research as it is the major signal transducer of both physioligcal, and perhaps more importantly, pathological angiogenesis, especially in cancerous tumors. VEGFR-2 is of critical importance to the body as exemplified by Shalaby ''et al.'' who demonstrated that VEGFR-2 gene knockout mice die at E8-8.5 due to lack of vasculogenesis.<ref>PMID:7596453</ref> The signal cascade initiated by binding VEGF to VEGFR is dependent upon specific sites of phosphorylation in the VEGFR structure and the interaction between these phosphorylated sites and other signaling molecules. | ||
| - | ==Structure of VEGFR-2 and Biology | + | ==Structure of VEGFR-2 and Biology== |
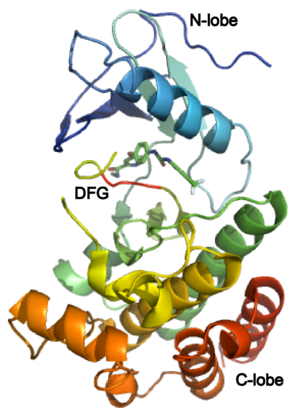
The structure of VEGFR-2 can been seen at the right. VEGF-A binds to the second and third extracellular Ig-like domains of VEGFR-2 with a 10-fold lower affinity than it does to the second Ig-like domain of VEGFR-1, despite the fact that VEGFR-2 is the principal mediator of several physiological effects on endothelial cells including proliferation, migration, and survival.<ref> PMID:9813036</ref> Binding of VEGF to the domains 2 and 3 of a VEGFR-2 monomer increases the probability that an additional VEGFR-2 binds the tethered ligand to form a dimmer. Once the two receptors are cross-linked, interactions between their membrane-proximal domain 7s stabilize the dimmer significantly. This dimerization and stabilization allows for precise positioning of the intracellular kinase domains, resulting in autophosphorylation and subsequent activation of the classical extracellular signal-regulated kinases (ERK) pathway.<ref>PMID:17293873</ref> | The structure of VEGFR-2 can been seen at the right. VEGF-A binds to the second and third extracellular Ig-like domains of VEGFR-2 with a 10-fold lower affinity than it does to the second Ig-like domain of VEGFR-1, despite the fact that VEGFR-2 is the principal mediator of several physiological effects on endothelial cells including proliferation, migration, and survival.<ref> PMID:9813036</ref> Binding of VEGF to the domains 2 and 3 of a VEGFR-2 monomer increases the probability that an additional VEGFR-2 binds the tethered ligand to form a dimmer. Once the two receptors are cross-linked, interactions between their membrane-proximal domain 7s stabilize the dimmer significantly. This dimerization and stabilization allows for precise positioning of the intracellular kinase domains, resulting in autophosphorylation and subsequent activation of the classical extracellular signal-regulated kinases (ERK) pathway.<ref>PMID:17293873</ref> | ||
| Line 18: | Line 19: | ||
<br /> | <br /> | ||
| - | ==Medical== | + | ==Medical significance== |
[[Image: Sorafenib.png|300px|left|thumb| [[Sorafenib]], anti VEGFR drug targeting the MAP Kinase pathway, marketed by Bayer for Renal and Liver [[Cancer]].]] | [[Image: Sorafenib.png|300px|left|thumb| [[Sorafenib]], anti VEGFR drug targeting the MAP Kinase pathway, marketed by Bayer for Renal and Liver [[Cancer]].]] | ||
{{Clear}} | {{Clear}} | ||
| Line 29: | Line 30: | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | + | *VEGFR-1 | |
| - | [[1rv6]] - hVEGFR-1 domain 2 + PlGF - human <br /> | + | **[[1rv6]] - hVEGFR-1 domain 2 + PlGF - human <br /> |
| - | [[1qsv]] - hVEGFR-1 VEGF-binding domain – NMR<br /> | + | **[[1qsv]] - hVEGFR-1 VEGF-binding domain – NMR<br /> |
| - | [[2xac]] – hVEGFR-1D2 + hVEGF-B<br /> | + | **[[2xac]] – hVEGFR-1D2 + hVEGF-B<br /> |
| - | [[3hng]] - hVEGFR-1 kinase domain + N-(4-Chlorophenyl)-2-((pyridin-4-ylmethyl)amino)benzamid <br /> | + | **[[3hng]] - hVEGFR-1 kinase domain + N-(4-Chlorophenyl)-2-((pyridin-4-ylmethyl)amino)benzamid <br /> |
| - | + | *VEGFR-2 | |
| - | [[3kvq]] - hVEGFR-2 extracellular domain 7<br /> | + | **[[3kvq]] - hVEGFR-2 extracellular domain 7<br /> |
| - | [[1vr2]] - hVEGFR-2 kinase domain (mutant)<br /> | + | **[[1vr2]] - hVEGFR-2 kinase domain (mutant)<br /> |
| - | [[2m59]] - hVEGFR-2 membrane domain – NMR<br /> | + | **[[2m59]] - hVEGFR-2 membrane domain – NMR<br /> |
| - | [[2x1w]], [[2x1x]] – hVEGFR-2 IG-like domains 2 & 3 +hVEGF-C<br /> | + | **[[2x1w]], [[2x1x]] – hVEGFR-2 IG-like domains 2 & 3 +hVEGF-C<br /> |
| - | [[3v2a]] - hVEGFR-2 + hVEGF-A<br /> | + | **[[3v2a]] - hVEGFR-2 + hVEGF-A<br /> |
| - | [[3v6b]] - hVEGFR-2 residues 132-548 + hVEGF-E<br /> | + | **[[3v6b]] - hVEGFR-2 residues 132-548 + hVEGF-E<br /> |
| - | [[3efl]] – hVEGFR-2 kinase domain (mutant)+ motesanib <br /> | + | **[[3efl]] – hVEGFR-2 kinase domain (mutant)+ motesanib <br /> |
| - | [[4ag8]], [[4agc]], [[4agd]], [[4asd]], [[4ase]], [[3vo3]] - hVEGFR-2 kinase domain (mutant) + renal cell carcinoma drug<br /> | + | **[[4ag8]], [[4agc]], [[4agd]], [[4asd]], [[4ase]], [[3vo3]] - hVEGFR-2 kinase domain (mutant) + renal cell carcinoma drug<br /> |
| - | [[3ewh]], [[3cjf]], [[3cjg]], [[3vhe]] - hVEGFR-2 kinase domain + pyrimidine derivative<br /> | + | **[[3ewh]], [[3cjf]], [[3cjg]], [[3vhe]] - hVEGFR-2 kinase domain + pyrimidine derivative<br /> |
| - | [[3c7q]] - hVEGFR-2 kinase domain + BIBF1120<br /> | + | **[[3c7q]] - hVEGFR-2 kinase domain + BIBF1120<br /> |
| - | [[3dtw]] - hVEGFR-2 kinase domain + benzisoxazole <br /> | + | **[[3dtw]] - hVEGFR-2 kinase domain + benzisoxazole <br /> |
| - | [[3cp9]], [[3cpb]], [[3cpc]], [[3be2]], [[3b8q]], [[3b8r]], [[2qu5]], [[2qu6]], [[2p2h]], [[2p2i]], [[1ywn]], [[1y6a]], [[1y6b]],[[3vid]], [[3vhk]] - hVEGFR-2 kinase domain + inhibitor<br /> | + | **[[3cp9]], [[3cpb]], [[3cpc]], [[3be2]], [[3b8q]], [[3b8r]], [[2qu5]], [[2qu6]], [[2p2h]], [[2p2i]], [[1ywn]], [[1y6a]], [[1y6b]],[[3vid]], [[3vhk]] - hVEGFR-2 kinase domain + inhibitor<br /> |
| - | [[2oh4]], [[2rl5]], [[2xir]], [[3u6j]], [[3vnt]] - hVEGFR-2 kinase domain (mutant)+ inhibitor<br /> | + | **[[2oh4]], [[2rl5]], [[2xir]], [[3u6j]], [[3vnt]] - hVEGFR-2 kinase domain (mutant)+ inhibitor<br /> |
| - | [[3s35]], [[3s36]], [[3s37]] - hVEGFR-2 extracellular domain 3 + Fab heavy+light chains | + | **[[3s35]], [[3s36]], [[3s37]] - hVEGFR-2 extracellular domain 3 + Fab heavy+light chains |
| - | + | *VEGFR-3 | |
| - | [[4bsj]] - hVEGFR-3 extracellular domain 4 & 5<br /> | + | **[[4bsj]] - hVEGFR-3 extracellular domain 4 & 5<br /> |
| - | [[4bsk]] - hVEGFR-3 extracellular domain 1 & 2 + VEGF-C<br /> | + | **[[4bsk]] - hVEGFR-3 extracellular domain 1 & 2 + VEGF-C<br /> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | *VEGF 165 receptor (neuropilin) see [[Neuropilin]] | ||
| + | }} | ||
==Additional Resources== | ==Additional Resources== | ||
Revision as of 10:10, 17 December 2014
| |||||||||||
Contents |
3D Structures of VEGFR
Updated on 17-December-2014
Additional Resources
For additional information, see: Cancer
See Also
References
- ↑ Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006 May;7(5):359-71. PMID:16633338 doi:10.1038/nrm1911
- ↑ Holmes K, Roberts OL, Thomas AM, Cross MJ. Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal. 2007 Oct;19(10):2003-12. Epub 2007 Jun 12. PMID:17658244 doi:10.1016/j.cellsig.2007.05.013
- ↑ Gallina P, Nohra G, Cioloca C, Meder JF, Roux FX. [Multiple cavernoma of delayed appearance] Neurochirurgie. 1994;40(5):322-5. PMID:7596453
- ↑ Shinkai A, Ito M, Anazawa H, Yamaguchi S, Shitara K, Shibuya M. Mapping of the sites involved in ligand association and dissociation at the extracellular domain of the kinase insert domain-containing receptor for vascular endothelial growth factor. J Biol Chem. 1998 Nov 20;273(47):31283-8. PMID:9813036
- ↑ Ruch C, Skiniotis G, Steinmetz MO, Walz T, Ballmer-Hofer K. Structure of a VEGF-VEGF receptor complex determined by electron microscopy. Nat Struct Mol Biol. 2007 Mar;14(3):249-50. Epub 2007 Feb 11. PMID:17293873 doi:10.1038/nsmb1202
- ↑ Shibuya M, Yamaguchi S, Yamane A, Ikeda T, Tojo A, Matsushime H, Sato M. Nucleotide sequence and expression of a novel human receptor-type tyrosine kinase gene (flt) closely related to the fms family. Oncogene. 1990 Apr;5(4):519-24. PMID:2158038
- ↑ Ji QS, Winnier GE, Niswender KD, Horstman D, Wisdom R, Magnuson MA, Carpenter G. Essential role of the tyrosine kinase substrate phospholipase C-gamma1 in mammalian growth and development. Proc Natl Acad Sci U S A. 1997 Apr 1;94(7):2999-3003. PMID:9096335
- ↑ Welsh M, Songyang Z, Frantz JD, Trub T, Reedquist KA, Karlsson T, Miyazaki M, Cantley LC, Band H, Shoelson SE. Stimulation through the T cell receptor leads to interactions between SHB and several signaling proteins. Oncogene. 1998 Feb 19;16(7):891-901. PMID:9484780 doi:10.1038/sj.onc.1201607
- ↑ Zeng H, Sanyal S, Mukhopadhyay D. Tyrosine residues 951 and 1059 of vascular endothelial growth factor receptor-2 (KDR) are essential for vascular permeability factor/vascular endothelial growth factor-induced endothelium migration and proliferation, respectively. J Biol Chem. 2001 Aug 31;276(35):32714-9. Epub 2001 Jul 2. PMID:11435426 doi:10.1074/jbc.M103130200
- ↑ Chen M, She H, Davis EM, Spicer CM, Kim L, Ren R, Le Beau MM, Li W. Identification of Nck family genes, chromosomal localization, expression, and signaling specificity. J Biol Chem. 1998 Sep 25;273(39):25171-8. PMID:9737977
- ↑ Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer. 2002 Oct;2(10):795-803. PMID:12360282 doi:10.1038/nrc909
- ↑ Rosa DD, Ismael G, Lago LD, Awada A. Molecular-targeted therapies: lessons from years of clinical development. Cancer Treat Rev. 2008 Feb;34(1):61-80. Epub 2007 Sep 10. PMID:17826917 doi:10.1016/j.ctrv.2007.07.019
- ↑ Jain RK. Normalizing tumor vasculature with anti-angiogenic therapy: a new paradigm for combination therapy. Nat Med. 2001 Sep;7(9):987-9. PMID:11533692 doi:10.1038/nm0901-987
- ↑ Rosa DD, Ismael G, Lago LD, Awada A. Molecular-targeted therapies: lessons from years of clinical development. Cancer Treat Rev. 2008 Feb;34(1):61-80. Epub 2007 Sep 10. PMID:17826917 doi:10.1016/j.ctrv.2007.07.019
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky, Wayne Decatur