1pgo
From Proteopedia
| Line 1: | Line 1: | ||
| - | [[Image:1pgo.jpg|left|200px]] | + | [[Image:1pgo.jpg|left|200px]] |
| - | + | ||
| - | '''CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM''' | + | {{Structure |
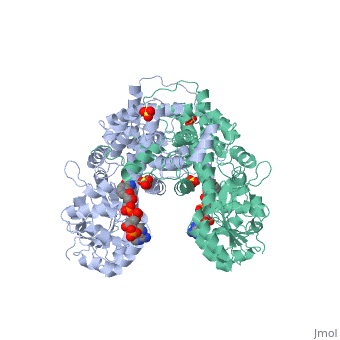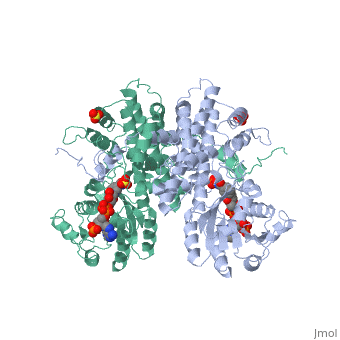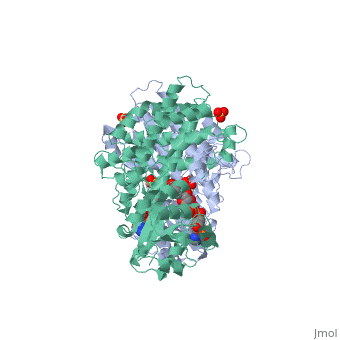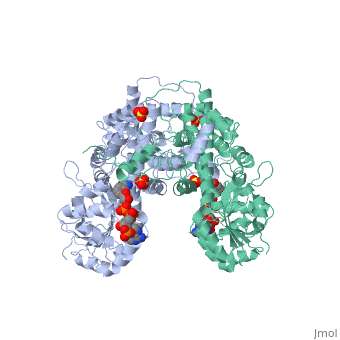
| + | |PDB= 1pgo |SIZE=350|CAPTION= <scene name='initialview01'>1pgo</scene>, resolution 2.5Å | ||
| + | |SITE= | ||
| + | |LIGAND= <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene> and <scene name='pdbligand=NDP:NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE'>NDP</scene> | ||
| + | |ACTIVITY= [http://en.wikipedia.org/wiki/Phosphogluconate_dehydrogenase_(decarboxylating) Phosphogluconate dehydrogenase (decarboxylating)], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.1.1.44 1.1.1.44] | ||
| + | |GENE= | ||
| + | }} | ||
| + | |||
| + | '''CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM''' | ||
| + | |||
==Overview== | ==Overview== | ||
| Line 7: | Line 16: | ||
==About this Structure== | ==About this Structure== | ||
| - | 1PGO is a [ | + | 1PGO is a [[Single protein]] structure of sequence from [http://en.wikipedia.org/wiki/Ovis_aries Ovis aries]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1PGO OCA]. |
==Reference== | ==Reference== | ||
| - | Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism., Adams MJ, Ellis GH, Gover S, Naylor CE, Phillips C, Structure. 1994 Jul 15;2(7):651-68. PMID:[http:// | + | Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism., Adams MJ, Ellis GH, Gover S, Naylor CE, Phillips C, Structure. 1994 Jul 15;2(7):651-68. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/7922042 7922042] |
[[Category: Ovis aries]] | [[Category: Ovis aries]] | ||
[[Category: Phosphogluconate dehydrogenase (decarboxylating)]] | [[Category: Phosphogluconate dehydrogenase (decarboxylating)]] | ||
| Line 21: | Line 30: | ||
[[Category: oxidoreductase (choh(d)-nadp+(a))]] | [[Category: oxidoreductase (choh(d)-nadp+(a))]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Mar 20 13:24:06 2008'' |
Revision as of 11:24, 20 March 2008
| |||||||
| , resolution 2.5Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | and | ||||||
| Activity: | Phosphogluconate dehydrogenase (decarboxylating), with EC number 1.1.1.44 | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM
Overview
BACKGROUND: The nicotinamide adenine dinucleotide phosphate (NADP)-dependent oxidative decarboxylase, 6-phosphogluconate dehydrogenase, is a major source of reduced coenzyme for synthesis. Enzymes later in the pentose phosphate pathway convert the reaction product, ribulose 5-phosphate, to ribose 5-phosphate. Crystallographic study of complexes with coenzyme and substrate explain the NADP dependence which determines the enzyme's metabolic role and support the proposed general base-general acid mechanism. RESULTS: The refined structures of binary coenzyme/analogue complexes show that Arg33 is ordered by binding the 2'-phosphate, and provides one face of the adenine site. The nicotinamide, while less tightly bound, is more extended when reduced than when oxidized. All substrate binding residues are conserved; the 3-hydroxyl of 6-phosphogluconate is hydrogen bonded to N zeta of Lys183 and the 3-hydrogen points towards the oxidized nicotinamide. The 6-phosphate replaces a tightly bound sulphate in the apo-enzyme. CONCLUSIONS: NADP specificity is achieved primarily by Arg33 which binds the 2'-phosphate but, in its absence, obscures the adenine pocket. The bound oxidized nicotinamide is syn; hydride transfer from bound substrate to the nicotinamide si- face is achieved with a small movement of the nicotinamide nucleotide. Lys183 may act as general base. A water bound to Gly130 in the coenzyme domain is the most likely acid required in decarboxylation. The dihydronicotinamide ring of NADPH competes for ligands with the 1-carboxyl of 6-phosphogluconate.
About this Structure
1PGO is a Single protein structure of sequence from Ovis aries. Full crystallographic information is available from OCA.
Reference
Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism., Adams MJ, Ellis GH, Gover S, Naylor CE, Phillips C, Structure. 1994 Jul 15;2(7):651-68. PMID:7922042
Page seeded by OCA on Thu Mar 20 13:24:06 2008