Sandbox Reserved 960
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
{{Sandbox_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | ==Crystal structure of the Antennal Specific Protein-1 from Apis mellifera (AmelASP1) with a serendipitous ligand at pH 5.5== | + | ==Crystal structure of the Antennal Specific Protein-1 from ''Apis mellifera'' (AmelASP1) with a serendipitous ligand at pH 5.5== |
<StructureSection load='3fe6' size='400' side='right' | <StructureSection load='3fe6' size='400' side='right' | ||
This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| Line 21: | Line 21: | ||
===Social relevance=== | ===Social relevance=== | ||
| - | The | + | The diets of workers bees and queen bee are strongly different and determinate diverse behaviors. |
Workers bee are fed with royal jelly for only three days after egg-laying whereas the queen bee eats royal jelly during her whole life. She controls the activity of each bees by chemical communication. | Workers bee are fed with royal jelly for only three days after egg-laying whereas the queen bee eats royal jelly during her whole life. She controls the activity of each bees by chemical communication. | ||
Actually, the queen bee is the only one able to produce 9-ODA - the main component of its pheromone which induces sexual or endocrine responses. This substance is sent to the workers bees which detect it through pheromone-binding proteins (PBPs). It is then transformed in 9-HDA and added in the royal jelly. This last substance is eaten by the queen. In turn, queen bee transforms 9-HDA into 9-ODA. | Actually, the queen bee is the only one able to produce 9-ODA - the main component of its pheromone which induces sexual or endocrine responses. This substance is sent to the workers bees which detect it through pheromone-binding proteins (PBPs). It is then transformed in 9-HDA and added in the royal jelly. This last substance is eaten by the queen. In turn, queen bee transforms 9-HDA into 9-ODA. | ||
| - | Thus, ASP1 is primordial to the internal | + | Thus, ASP1 is primordial to the internal pheromone’s transport cycle in the hive. By binding the component of queen bee pheromone, bees express and transmit essential behaviour within the hive. Indeed, 9-ODA is responsible, among others, of preventing workers bees’ ovarian development. |
===Location in the antenna and transport of pheromones=== | ===Location in the antenna and transport of pheromones=== | ||
| Line 31: | Line 31: | ||
== Structure == | == Structure == | ||
| - | |||
| - | [[Image: 3fe6_cartoon.jpg|250px|left|thumb|'''Fig.1''' Ribbon colored representation]] | ||
| - | |||
| - | |||
=== Domains and family === | === Domains and family === | ||
The C terminal(scene) domain of this molecule presents a characteristic PBP-GOP domain. While this protein is composed of 144 residues the PBP domain begins at the 25th residue. | The C terminal(scene) domain of this molecule presents a characteristic PBP-GOP domain. While this protein is composed of 144 residues the PBP domain begins at the 25th residue. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== Key residues === | === Key residues === | ||
| Line 63: | Line 49: | ||
AmelASP1 presents <scene name='60/604479/Disulfide_bonds/1'> three disulfide bridges</scene> which are greatly enhancing its structure’s rigidity by linking four of the helices together. The six cysteines and their interval spacing are the most striking features shared by proteins belonging to the PBP family. | AmelASP1 presents <scene name='60/604479/Disulfide_bonds/1'> three disulfide bridges</scene> which are greatly enhancing its structure’s rigidity by linking four of the helices together. The six cysteines and their interval spacing are the most striking features shared by proteins belonging to the PBP family. | ||
| - | The <scene name='60/604479/1st_disulfide_bridge/1'>first disulfide bridge</scene> is established between <scene name='60/604479/H1/2'>H1</scene> and <scene name='60/604479/H3/2'>H3</scene> through | + | The <scene name='60/604479/1st_disulfide_bridge/1'>first disulfide bridge</scene> is established between <scene name='60/604479/H1/2'>H1</scene> and <scene name='60/604479/H3/2'>H3</scene> through Cysteines 20 and 51. <scene name='60/604479/2nd_disulfide_bridge/2'>An other disulfide bridge</scene> links <scene name='60/604479/H3/2'>H3</scene> and <scene name='60/604479/H6/1'>H6</scene> through Cys 47 and 98, and the <scene name='60/604479/3rd_disulfide_bridge/1'>third disulfide bridge</scene> connects <scene name='60/604479/H5/1'>H5</scene> and <scene name='60/604479/H6/1'>H6</scene> thanks to Cys 89 and Cys 107. |
Furthermore, non covalent bonds also play an important role. | Furthermore, non covalent bonds also play an important role. | ||
| - | Indeed, at pH 5.5, among the numerous other, two | + | Indeed, at pH 5.5, among the numerous other, two hydrogen bonds are particularly noticeable because of their importance in the formation of the loop stabilizing H4. <scene name='60/604479/Hydrogene_bond_1/2'>One</scene> is established by Asp 66 and Leu 58 whereas <scene name='60/604479/Hydrogene_bond_2/1'>the second</scene> is formed between Asp 60 and Ala 63. |
=== Cavity === | === Cavity === | ||
| Line 76: | Line 62: | ||
In fact, depending of the pH level, Asp35 bend the C terminal domain against the cavity. | In fact, depending of the pH level, Asp35 bend the C terminal domain against the cavity. | ||
At pH 5.5, Asp35 is protonated and C terminal domain isn’t bend against the cavity. While ASP1 is a monomere at acid pH, it can dimerize at neutral and basic pH. | At pH 5.5, Asp35 is protonated and C terminal domain isn’t bend against the cavity. While ASP1 is a monomere at acid pH, it can dimerize at neutral and basic pH. | ||
| - | |||
== Ligands == | == Ligands == | ||
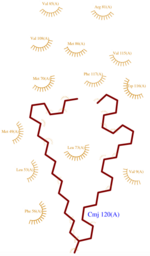
| - | + | [[Image:CMJ_Ligplot.png|150px|right|thumb|'''Fig.2''' CMJ Ligplot]] | |
In order to determine this protein’s structure, several ligands has been used at pH 5.5 because this low pH fits with the natural medium of this protein in the bee antenna. | In order to determine this protein’s structure, several ligands has been used at pH 5.5 because this low pH fits with the natural medium of this protein in the bee antenna. | ||
| + | The three ligands used to characterize and purify AmelASP1 are : | ||
| + | *<scene name='60/604479/Cmj/3'>CMJ</scene> also known as (20s)-20-Methyldotetracontane, is a serendipitous ligand. This term signify that the purification of this molecule was completely fortuitous. It is a big unsaturated mono-methyl branched carbone chain with formula C43H88. This ligand fits in the hydrophobic cavity of AmelASP1 thanks to several interactions with <scene name='60/604479/Cmj_binding_residues/2'>specific residues.</scene> | ||
| - | The three ligands used to caracterize and purify AmelASP1 are : | ||
| - | *<scene name='60/604479/Cmj/3'>CMJ</scene> also known as (20s)-20-Methyldotetracontane, is a serendipitous ligand. This term signify that the purification of this molecule was completly fortuitous. It is a big unsatured mono-methyl branched carbone chain with formula C43H88. This ligand fits in the hydrophobic cavity of AmelASP1 thanks to several interactions with <scene name='60/604479/Cmj_binding_residues/2'>specific residues.</scene> | ||
| - | [[Image:CMJ_Ligplot.png|150px|right|thumb|'''Fig.2''' CMJ Ligplot]] | ||
| - | *<scene name='60/604479/Gol/1'>Glycerol</scene> (C3H8O3) also known as GOL, is a ligand used for cryoprotection during the purification process of the protein. It is supposedly helping the main ligand to reach its binding site. To do so, GOL links to<scene name='60/604479/Gol_binding_residues/1'>Asn 41 and Tyr 102.</scene> | ||
| - | [[Image:GOL_Ligplot.png|200px|left|thumb|'''Fig.3''' GOL Ligplot]] | ||
| - | *Chloride ion facilitates the binding of other ligands to the protein. Its abundance around ASP1 depends of the condition. | ||
| - | <scene name='60/604479/Gol_binding_residues/1'>GOL binding residues</scene> | ||
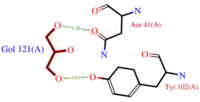
| + | *<scene name='60/604479/Gol/1'>Glycerol</scene> (C3H8O3) also known as GOL, is a ligand used for cryoprotection during the purification process of the protein. It is supposedly helping the main ligand to reach its binding site. To do so, GOL links to<scene name='60/604479/Gol_binding_residues/1'> Asn 41 and Tyr 102.</scene> | ||
| + | [[Image:GOL_Ligplot.png|200px|left|thumb|'''Fig.3''' GOL Ligplot]] | ||
| - | [[Image:Cl_Ligplot.png|right|thumb|'''Fig.4''' Cl Ligplot]] | ||
| - | <scene name='60/604479/Cl/1'>Cl Ligand</scene> | ||
| - | <scene name='60/604479/Cl_binding_residue/1'>Cl binding residue</scene> | ||
| - | === Natural ligands === | ||
| Line 118: | Line 96: | ||
| - | <scene name='60/604479/Hydrophobic_residues/1'>hydrophobic residues</scene> | ||
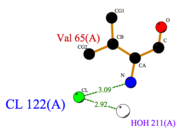
| - | <scene name='60/604479/ | + | *<scene name='60/604479/Cl/1'>Chloride ion</scene> facilitates the binding of other ligands to the protein. Its abundance around ASP1 varies according to changing pH conditions. At pH 5.5, <scene name='60/604479/Cl_binding_residue/1'>Val 65</scene> is the only AA able to fix a Cl ion. |
| + | [[Image:Cl_Ligplot.png|right|thumb|'''Fig.4''' Cl Ligplot]] | ||
| - | <scene name='60/604479/Acidic_residues/1'>acidic residues</scene> | ||
| - | == Related structures == | ||
| - | These structures shown below representing the same protein with variable ligand and pH emphasize and illustrate the binding versatility of PBP. | ||
| - | *[[3fe8]] The same protein in complex with a serendipitous ligand soaked at pH 4.0 <br /> | ||
| - | *[[3fe9]] The same protein in complex with a serendipitous ligand soaked at pH 7.0 <br /> | ||
| - | *[[3cdn]] The same protein in apo form soaked at pH 4.0 <br /> | ||
| - | *[[2h8v]] The same protein in apo form at pH 5.5 <br /> | ||
| - | *[[3cz2]] The same protein in apo form at pH 7.0 <br /> | ||
| - | *[[3bfa]] The same protein in complex with the QMP at pH 5.5 <br /> | ||
| - | *[[3bfb]] The same protein in complex with the 9-ODA at pH 5.5 <br /> | ||
| - | *[[3bfh]] The same protein in complex with the HDOA at pH 5.5 <br /> | ||
| - | *[[3bjh]] The same protein in complex with the nBBSA at pH 5.5 <br /> | ||
| - | *[[3cyz]] The same protein in complex with the 9-ODA at pH 7.0 <br /> | ||
| - | *[[3cz0]] The same protein in complex with the QMP at pH 7.0 <br /> | ||
| - | *[[3cz1]] The same protein in complex with the nBBSA at pH 7.0 <br /> | ||
| - | *[[3cab]] The same protein in complex with the nBBSA soaked at pH 7.0 <br /> | ||
| Line 151: | Line 113: | ||
| - | == References for further information on the pheromone binding protein from Apis mellifera == | + | |
| + | However, in natural conditions, binding ligands are the pheromones secreted by the queen such as 9-ODA. | ||
| + | |||
| + | == Related structures == | ||
| + | The structures shown below representing AmelASP1 with variable ligands and pH, emphasize and illustrate the binding versatility of Pheromones Binding Proteins. | ||
| + | The same PBP and their ligands can be crystalized either in apo (without ligand) or holo states (with ligand). | ||
| + | *In the apo state, the C terminus part of the protein obstructs the binding site. | ||
| + | **[[3cdn]] The same protein in apo form soaked at pH 4.0 <br /> | ||
| + | **[[2h8v]] The same protein in apo form at pH 5.5 <br /> | ||
| + | **[[3cz2]] The same protein in apo form at pH 7.0 <br /> | ||
| + | |||
| + | *In the holo state, depending of the ligand, the volume of the cavity changes. Likewise, presence of ligand and favorable pH, induce C terminus conformation changes. | ||
| + | **[[3fe8]] The same protein in complex with a serendipitous ligand soaked at pH 4.0 <br /> | ||
| + | **[[3fe9]] The same protein in complex with a serendipitous ligand soaked at pH 7.0 <br /> | ||
| + | **[[3bfa]] The same protein in complex with the QMP at pH 5.5 <br /> | ||
| + | **[[3bfb]] The same protein in complex with the 9-ODA at pH 5.5 <br /> | ||
| + | **[[3bfh]] The same protein in complex with the HDOA at pH 5.5 <br /> | ||
| + | **[[3bjh]] The same protein in complex with the nBBSA at pH 5.5 <br /> | ||
| + | **[[3cyz]] The same protein in complex with the 9-ODA at pH 7.0 <br /> | ||
| + | **[[3cz0]] The same protein in complex with the QMP at pH 7.0 <br /> | ||
| + | **[[3cz1]] The same protein in complex with the nBBSA at pH 7.0 <br /> | ||
| + | **[[3cab]] The same protein in complex with the nBBSA soaked at pH 7.0 <br /> | ||
| + | |||
| + | </StructureSection> | ||
| + | == Contributors == | ||
| + | Sophie Morin & Mathias Buytaert | ||
| + | == References for further information on the pheromone binding protein from ''Apis mellifera'' == | ||
<references/> | <references/> | ||
Revision as of 22:14, 23 December 2014
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
Crystal structure of the Antennal Specific Protein-1 from Apis mellifera (AmelASP1) with a serendipitous ligand at pH 5.5
| |||||||||||
Contributors
Sophie Morin & Mathias Buytaert
References for further information on the pheromone binding protein from Apis mellifera
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Tegoni M, Cambillau C. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J Mol Biol. 2008 Jun 27;380(1):158-69. Epub 2008 Apr 27. PMID:18508083 doi:10.1016/j.jmb.2008.04.048
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Campanacci V, Tegoni M, Cambillau C. Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release. J Mol Biol. 2009 Jul 31;390(5):981-90. Epub 2009 May 28. PMID:19481550 doi:10.1016/j.jmb.2009.05.067
- ↑ Han L, Zhang YJ, Zhang L, Cui X, Yu J, Zhang Z, Liu MS. Operating mechanism and molecular dynamics of pheromone-binding protein ASP1 as influenced by pH. PLoS One. 2014 Oct 22;9(10):e110565. doi: 10.1371/journal.pone.0110565., eCollection 2014. PMID:25337796 doi:http://dx.doi.org/10.1371/journal.pone.0110565
- ↑ Lartigue A, Gruez A, Briand L, Blon F, Bezirard V, Walsh M, Pernollet JC, Tegoni M, Cambillau C. Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L. J Biol Chem. 2004 Feb 6;279(6):4459-64. Epub 2003 Oct 31. PMID:14594955 doi:10.1074/jbc.M311212200
- ↑ http://www.genome.jp/dbget-bin/www_bget?pdb:3FE6