Sandbox Reserved 960
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Biological function== | ==Biological function== | ||
| Line 56: | Line 56: | ||
=== Cavity === | === Cavity === | ||
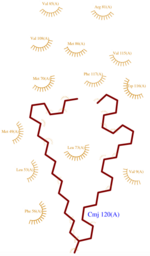
The dynamic structure of the protein is responsible of the ligand’s binding by adjustment of position. The successful delivery of the effector to the receptor relies on this property. The ligand binding pocket consists in a cavity formed by the helices H2, H4 and H5 (scene), arranged in a globular shape which leads to a clear separation of the ligand from the {{Template:ColorKey_Polar}} environment. | The dynamic structure of the protein is responsible of the ligand’s binding by adjustment of position. The successful delivery of the effector to the receptor relies on this property. The ligand binding pocket consists in a cavity formed by the helices H2, H4 and H5 (scene), arranged in a globular shape which leads to a clear separation of the ligand from the {{Template:ColorKey_Polar}} environment. | ||
| - | The top of the cavity is not closed and can establish contacts with the solvent. The cavity is prone to accept ligand such as 9-ODA because of its specific composition. Indeed, cavity's components are mainly <scene name='60/604479/hydrophobic_residues/2'>hydrophobic and aromatic</scene>.They consequently interact with the ligand's {{Template:ColorKey_Hydrophobic}} carbon chain and are localized on the internal face of the helix.Thus, it implies that these residues respect a regular distance pattern in the primary structure of the AmelASP1. | + | The top of the cavity is not closed and can establish contacts with the solvent. The cavity is prone to accept ligand such as 9-ODA because of its specific composition. Indeed, cavity's components are mainly <scene name='60/604479/hydrophobic_residues/2'>hydrophobic and aromatic</scene> <ref>PMID: 14594955</ref>.They consequently interact with the ligand's {{Template:ColorKey_Hydrophobic}} carbon chain and are localized on the internal face of the helix.Thus, it implies that these residues respect a regular distance pattern in the primary structure of the AmelASP1. |
=== pH influence === | === pH influence === | ||
| - | pH affects the flexibility of ASP1 because it induces a different protonation state of the <scene name='60/604479/Ionizable_residues/1'>ionizable residues</scene>. Protonated residues induce micro-environnment changes which propagate all along the protein. Consequently, ASP1 is no longer able to interact with its ligands even if ionizable residues are distant from the cavity. | + | pH affects the flexibility of ASP1 because it induces a different protonation state of the <scene name='60/604479/Ionizable_residues/1'>ionizable residues</scene>. Protonated residues induce micro-environnment changes which propagate all along the protein. Consequently, ASP1 is no longer able to interact with its ligands even if ionizable residues are distant from the cavity. <ref>PMID: 19481550</ref> |
In fact, depending of the pH level, Asp35 bend the C terminal domain against the cavity. | In fact, depending of the pH level, Asp35 bend the C terminal domain against the cavity. | ||
| - | At pH 5.5, <scene name='60/604479/C-term_asp35/1'>Asp 35</scene> is protonated and C terminal domain isn’t bend against the cavity. While ASP1 is a monomere at acid pH, it can dimerize at neutral and basic pH. | + | At pH 5.5, <scene name='60/604479/C-term_asp35/1'>Asp 35</scene> is protonated and C terminal domain isn’t bend against the cavity. While ASP1 is a monomere at acid pH, it can dimerize at neutral and basic pH. <ref>PMID: 25337796</ref> |
== Ligands == | == Ligands == | ||
Revision as of 09:10, 24 December 2014
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
Crystal structure of the Antennal Specific Protein-1 from Apis mellifera (AmelASP1) with a serendipitous ligand at pH 5.5
| |||||||||||
Contributors
Sophie Morin & Mathias Buytaert
References for further information on the pheromone binding protein from Apis mellifera
- ↑ http://www.genome.jp/dbget-bin/www_bget?pdb:3FE6
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Tegoni M, Cambillau C. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J Mol Biol. 2008 Jun 27;380(1):158-69. Epub 2008 Apr 27. PMID:18508083 doi:10.1016/j.jmb.2008.04.048
- ↑ Lartigue A, Gruez A, Briand L, Blon F, Bezirard V, Walsh M, Pernollet JC, Tegoni M, Cambillau C. Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L. J Biol Chem. 2004 Feb 6;279(6):4459-64. Epub 2003 Oct 31. PMID:14594955 doi:10.1074/jbc.M311212200
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Campanacci V, Tegoni M, Cambillau C. Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release. J Mol Biol. 2009 Jul 31;390(5):981-90. Epub 2009 May 28. PMID:19481550 doi:10.1016/j.jmb.2009.05.067
- ↑ Han L, Zhang YJ, Zhang L, Cui X, Yu J, Zhang Z, Liu MS. Operating mechanism and molecular dynamics of pheromone-binding protein ASP1 as influenced by pH. PLoS One. 2014 Oct 22;9(10):e110565. doi: 10.1371/journal.pone.0110565., eCollection 2014. PMID:25337796 doi:http://dx.doi.org/10.1371/journal.pone.0110565