Sandbox Reserved 960
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
{{Sandbox_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | + | =Crystal structure of the Antennal Specific Protein-1 from ''Apis mellifera'' (AmelASP1) with a serendipitous ligand at pH 5.5= | |
<StructureSection load='3fe6' size='400' side='right' | <StructureSection load='3fe6' size='400' side='right' | ||
This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| Line 37: | Line 37: | ||
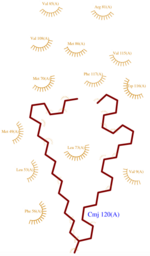
<scene name='60/604479/H1/2'>H1</scene> has a break in the hydrogen-bonding pattern of its structure, forming tight substitute hydrogen bonds with water molecules. Thus, it results in a <scene name='60/604479/Kink/1'>kink</scene> (at residue Ala 14) induced by a disruption in the helical conformation, due to hydrogen bonds with water molecules.<ref>PMID: 18508083</ref> | <scene name='60/604479/H1/2'>H1</scene> has a break in the hydrogen-bonding pattern of its structure, forming tight substitute hydrogen bonds with water molecules. Thus, it results in a <scene name='60/604479/Kink/1'>kink</scene> (at residue Ala 14) induced by a disruption in the helical conformation, due to hydrogen bonds with water molecules.<ref>PMID: 18508083</ref> | ||
| - | |||
=== Components implicated in the structure rigidity === | === Components implicated in the structure rigidity === | ||
| Line 57: | Line 56: | ||
== Ligands == | == Ligands == | ||
| + | [[Image:CMJ_Ligplot.png|150px|right|thumb|'''Fig.2''' CMJ Ligplot]] | ||
In order to determine this {{Template:ColorKey Composition Protein}}’s structure, several {{Template:ColorKey Composition Ligand}} has been used at pH 5.5 because this low pH fits with its natural medium in the bee antenna. | In order to determine this {{Template:ColorKey Composition Protein}}’s structure, several {{Template:ColorKey Composition Ligand}} has been used at pH 5.5 because this low pH fits with its natural medium in the bee antenna. | ||
| - | + | ||
The three ligands used to characterize and purify AmelASP1 are : | The three ligands used to characterize and purify AmelASP1 are : | ||
*<scene name='60/604479/Cmj/3'>CMJ</scene> also known as (20s)-20-Methyldotetracontane, is a serendipitous ligand. This term signify that the purification of this molecule was completely fortuitous. It is a big unsaturated mono-methyl branched carbone chain with formula C43H88. This ligand fits in the {{Template:ColorKey_Hydrophobic}} cavity of AmelASP1 thanks to several interactions with <scene name='60/604479/Cmj_binding_residues/2'>specific residues.</scene> | *<scene name='60/604479/Cmj/3'>CMJ</scene> also known as (20s)-20-Methyldotetracontane, is a serendipitous ligand. This term signify that the purification of this molecule was completely fortuitous. It is a big unsaturated mono-methyl branched carbone chain with formula C43H88. This ligand fits in the {{Template:ColorKey_Hydrophobic}} cavity of AmelASP1 thanks to several interactions with <scene name='60/604479/Cmj_binding_residues/2'>specific residues.</scene> | ||
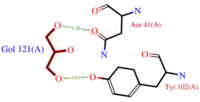
| + | *[[Image:GOL_Ligplot.png|200px|left|thumb|'''Fig.3''' GOL Ligplot]]<scene name='60/604479/Gol/1'>Glycerol</scene> (C3H8O3) also known as GOL, is a ligand used for cryoprotection during the purification process of the protein. It is supposedly helping the main ligand to reach its binding site. To do so, GOL links to<scene name='60/604479/Gol_binding_residues/1'> Asn 41 and Tyr 102.</scene> | ||
| + | [[Image:Cl_Ligplot.png|right|thumb|'''Fig.4''' Cl Ligplot]] | ||
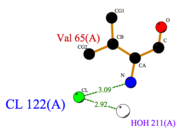
| - | + | *<scene name='60/604479/Cl/1'>Chloride ion</scene> facilitates the binding of other ligands to the protein. Its abundance around ASP1 varies according to changing pH conditions. At pH 5.5, <scene name='60/604479/Cl_binding_residue/1'>Val 65</scene> is the only amino acid able to fix a chloride ion. | |
| - | + | ||
| - | + | ||
| - | *<scene name='60/604479/Cl/1'>Chloride ion</scene> facilitates the binding of other ligands to the protein. | + | |
However, in natural conditions, binding ligands are the pheromones secreted by the queen such as 9-ODA. | However, in natural conditions, binding ligands are the pheromones secreted by the queen such as 9-ODA. | ||
Revision as of 09:24, 24 December 2014
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
Crystal structure of the Antennal Specific Protein-1 from Apis mellifera (AmelASP1) with a serendipitous ligand at pH 5.5
| |||||||||||
Contributors
Sophie Morin & Mathias Buytaert
Updated on 24-December-2014
References for further information on the pheromone binding protein from Apis mellifera
- ↑ http://www.genome.jp/dbget-bin/www_bget?pdb:3FE6
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Tegoni M, Cambillau C. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J Mol Biol. 2008 Jun 27;380(1):158-69. Epub 2008 Apr 27. PMID:18508083 doi:10.1016/j.jmb.2008.04.048
- ↑ Lartigue A, Gruez A, Briand L, Blon F, Bezirard V, Walsh M, Pernollet JC, Tegoni M, Cambillau C. Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L. J Biol Chem. 2004 Feb 6;279(6):4459-64. Epub 2003 Oct 31. PMID:14594955 doi:10.1074/jbc.M311212200
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Campanacci V, Tegoni M, Cambillau C. Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release. J Mol Biol. 2009 Jul 31;390(5):981-90. Epub 2009 May 28. PMID:19481550 doi:10.1016/j.jmb.2009.05.067
- ↑ Han L, Zhang YJ, Zhang L, Cui X, Yu J, Zhang Z, Liu MS. Operating mechanism and molecular dynamics of pheromone-binding protein ASP1 as influenced by pH. PLoS One. 2014 Oct 22;9(10):e110565. doi: 10.1371/journal.pone.0110565., eCollection 2014. PMID:25337796 doi:http://dx.doi.org/10.1371/journal.pone.0110565