This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
3m9s
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
== Structural highlights == | == Structural highlights == | ||
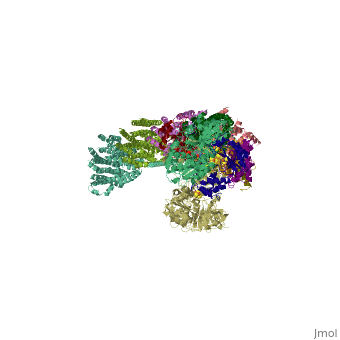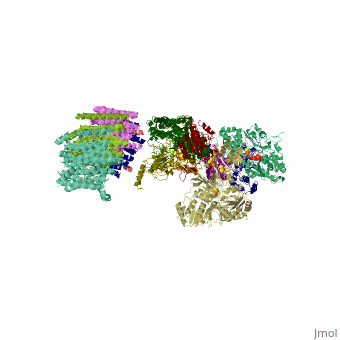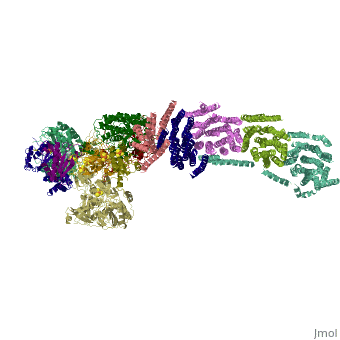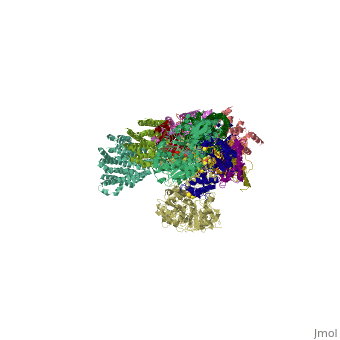
<table><tr><td colspan='2'>[[3m9s]] is a 26 chain structure with sequence from [http://en.wikipedia.org/wiki/Thermus_thermophilus Thermus thermophilus]. The December 2011 RCSB PDB [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Complex I'' by David Goodsell is [http://dx.doi.org/10.2210/rcsb_pdb/mom_2011_12 10.2210/rcsb_pdb/mom_2011_12]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3M9S OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3M9S FirstGlance]. <br> | <table><tr><td colspan='2'>[[3m9s]] is a 26 chain structure with sequence from [http://en.wikipedia.org/wiki/Thermus_thermophilus Thermus thermophilus]. The December 2011 RCSB PDB [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Complex I'' by David Goodsell is [http://dx.doi.org/10.2210/rcsb_pdb/mom_2011_12 10.2210/rcsb_pdb/mom_2011_12]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3M9S OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3M9S FirstGlance]. <br> | ||
| - | </td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=FES:FE2/S2+(INORGANIC)+CLUSTER'>FES</scene>, <scene name='pdbligand=FMN:FLAVIN+MONONUCLEOTIDE'>FMN</scene>, <scene name='pdbligand=SF4:IRON/SULFUR+CLUSTER'>SF4</scene>< | + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=FES:FE2/S2+(INORGANIC)+CLUSTER'>FES</scene>, <scene name='pdbligand=FMN:FLAVIN+MONONUCLEOTIDE'>FMN</scene>, <scene name='pdbligand=SF4:IRON/SULFUR+CLUSTER'>SF4</scene></td></tr> |
| - | <tr><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=UNK:UNKNOWN'>UNK</scene></td></tr> | + | <tr id='NonStdRes'><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=UNK:UNKNOWN'>UNK</scene></td></tr> |
| - | <tr><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3m9c|3m9c]]</td></tr> | + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3m9c|3m9c]]</td></tr> |
| - | <tr><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/NADH_dehydrogenase_(quinone) NADH dehydrogenase (quinone)], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.6.99.5 1.6.99.5] </span></td></tr> | + | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/NADH_dehydrogenase_(quinone) NADH dehydrogenase (quinone)], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.6.99.5 1.6.99.5] </span></td></tr> |
| - | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3m9s FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3m9s OCA], [http://www.rcsb.org/pdb/explore.do?structureId=3m9s RCSB], [http://www.ebi.ac.uk/pdbsum/3m9s PDBsum]</span></td></tr> | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3m9s FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3m9s OCA], [http://www.rcsb.org/pdb/explore.do?structureId=3m9s RCSB], [http://www.ebi.ac.uk/pdbsum/3m9s PDBsum]</span></td></tr> |
| - | <table> | + | </table> |
| + | == Function == | ||
| + | [[http://www.uniprot.org/uniprot/NQO6_THET8 NQO6_THET8]] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP.[HAMAP-Rule:MF_01356] [[http://www.uniprot.org/uniprot/NQO3_THET8 NQO3_THET8]] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP. [[http://www.uniprot.org/uniprot/NQO9_THET8 NQO9_THET8]] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP. The role of the nqo9 subunit appears to provide a 'connecting chain' of two clusters between cluster N5 and the terminal cluster N2, and to stabilize the structure of the complex by interacting with other subunits.[HAMAP-Rule:MF_01351] [[http://www.uniprot.org/uniprot/NQO2_THET8 NQO2_THET8]] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP. [[http://www.uniprot.org/uniprot/NQO15_THET8 NQO15_THET8]] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP. The nqo15 subunit has probably a role in complex stabilization, and may be also involved in the storage of iron for iron-sulfur cluster regeneration in the complex.<ref>PMID:16469879</ref> [[http://www.uniprot.org/uniprot/NQO1_THET8 NQO1_THET8]] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP. The nqo1 subunit contains the NADH-binding site and the primary electron acceptor FMN. [[http://www.uniprot.org/uniprot/NQO4_THET8 NQO4_THET8]] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP. The nqo4 subunit may contain the quinone-binding site.[HAMAP-Rule:MF_01358] [[http://www.uniprot.org/uniprot/NQO5_THET8 NQO5_THET8]] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP. The nqo5 subunit may be involved in the stabilization of the complex.[HAMAP-Rule:MF_01357] | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 25: | Line 27: | ||
The architecture of respiratory complex I.,Efremov RG, Baradaran R, Sazanov LA Nature. 2010 May 27;465(7297):441-5. PMID:20505720<ref>PMID:20505720</ref> | The architecture of respiratory complex I.,Efremov RG, Baradaran R, Sazanov LA Nature. 2010 May 27;465(7297):441-5. PMID:20505720<ref>PMID:20505720</ref> | ||
| - | From | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> |
</div> | </div> | ||
== References == | == References == | ||
| Line 34: | Line 36: | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
[[Category: Thermus thermophilus]] | [[Category: Thermus thermophilus]] | ||
| - | [[Category: Baradaran, R | + | [[Category: Baradaran, R]] |
| - | [[Category: Efremov, R G | + | [[Category: Efremov, R G]] |
| - | [[Category: Sazanov, L A | + | [[Category: Sazanov, L A]] |
[[Category: Complex i]] | [[Category: Complex i]] | ||
[[Category: Electron transport]] | [[Category: Electron transport]] | ||
Revision as of 04:56, 25 December 2014
Crystal structure of respiratory complex I from Thermus thermophilus
| |||||||||||