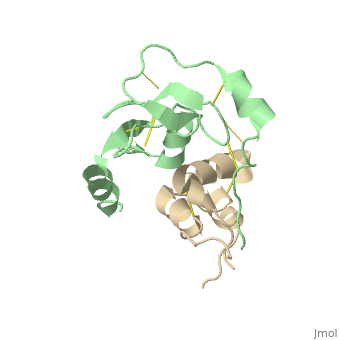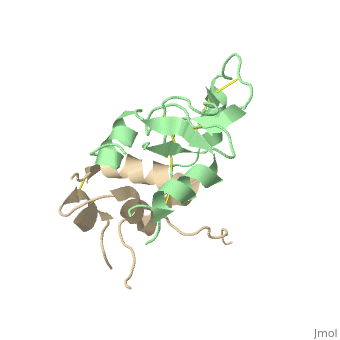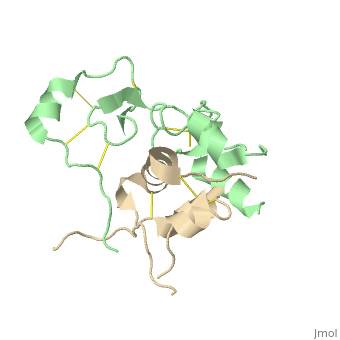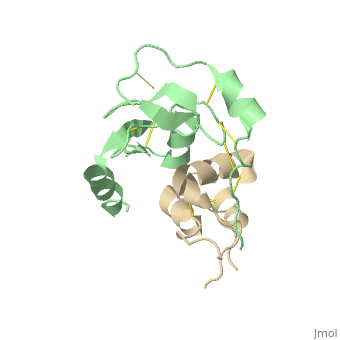2dsp
From Proteopedia
| Line 1: | Line 1: | ||
| - | [[Image:2dsp.gif|left|200px]] | + | [[Image:2dsp.gif|left|200px]] |
| - | + | ||
| - | '''Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins''' | + | {{Structure |
| + | |PDB= 2dsp |SIZE=350|CAPTION= <scene name='initialview01'>2dsp</scene>, resolution 2.50Å | ||
| + | |SITE= | ||
| + | |LIGAND= | ||
| + | |ACTIVITY= | ||
| + | |GENE= | ||
| + | }} | ||
| + | |||
| + | '''Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins''' | ||
| + | |||
==Overview== | ==Overview== | ||
| Line 10: | Line 19: | ||
==About this Structure== | ==About this Structure== | ||
| - | 2DSP is a [ | + | 2DSP is a [[Protein complex]] structure of sequences from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2DSP OCA]. |
==Reference== | ==Reference== | ||
| - | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins., Sitar T, Popowicz GM, Siwanowicz I, Huber R, Holak TA, Proc Natl Acad Sci U S A. 2006 Aug 29;103(35):13028-33. Epub 2006 Aug 21. PMID:[http:// | + | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins., Sitar T, Popowicz GM, Siwanowicz I, Huber R, Holak TA, Proc Natl Acad Sci U S A. 2006 Aug 29;103(35):13028-33. Epub 2006 Aug 21. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/16924115 16924115] |
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Protein complex]] | [[Category: Protein complex]] | ||
| Line 25: | Line 34: | ||
[[Category: insulin]] | [[Category: insulin]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Mar 20 16:31:03 2008'' |
Revision as of 14:31, 20 March 2008
| |||||||
| , resolution 2.50Å | |||||||
|---|---|---|---|---|---|---|---|
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins
Contents |
Overview
Insulin-like growth factor-binding proteins (IGFBPs) control bioavailability, activity, and distribution of insulin-like growth factor (IGF)1 and -2 through high-affinity IGFBP/IGF complexes. IGF-binding sites are found on N- and C-terminal fragments of IGFBPs, the two conserved domains of IGFBPs. The relative contributions of these domains to IGFBP/IGF complexation has been difficult to analyze, in part, because of the lack of appropriate three-dimensional structures. To analyze the effects of N- and C-terminal domain interactions, we determined several x-ray structures: first, of a ternary complex of N- and C-terminal domain fragments of IGFBP4 and IGF1 and second, of a "hybrid" ternary complex using the C-terminal domain fragment of IGFBP1 instead of IGFBP4. We also solved the binary complex of the N-terminal domains of IGFBP4 and IGF1, again to analyze C- and N-terminal domain interactions by comparison with the ternary complexes. The structures reveal the mechanisms of IGF signaling regulation via IGFBP binding. This finding supports research into the design of IGFBP variants as therapeutic IGF inhibitors for diseases of IGF disregulation. In IGFBP4, residues 1-38 form a rigid disulphide bond ladder-like structure, and the first five N-terminal residues bind to IGF and partially mask IGF residues responsible for the type 1 IGF receptor binding. A high-affinity IGF1-binding site is located in a globular structure between residues 39 and 82. Although the C-terminal domains do not form stable binary complexes with either IGF1 or the N-terminal domain of IGFBP4, in the ternary complex, the C-terminal domain contacts both and contributes to blocking of the IGF1 receptor-binding region of IGF1.
Disease
Known disease associated with this structure: Growth retardation with deafness and mental retardation due to IGF1 deficiency OMIM:[147440]
About this Structure
2DSP is a Protein complex structure of sequences from Homo sapiens. Full crystallographic information is available from OCA.
Reference
Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins., Sitar T, Popowicz GM, Siwanowicz I, Huber R, Holak TA, Proc Natl Acad Sci U S A. 2006 Aug 29;103(35):13028-33. Epub 2006 Aug 21. PMID:16924115
Page seeded by OCA on Thu Mar 20 16:31:03 2008
Categories: Homo sapiens | Protein complex | Holak, T A. | Huber, R. | Popowicz, G M. | Sitar, T. | Siwanowicz, I. | Igf | Igfbp | Insulin